|
Symbol Name ID |
Mgat5b
mannoside acetylglucosaminyltransferase 5, isoenzyme B MGI:3606200 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
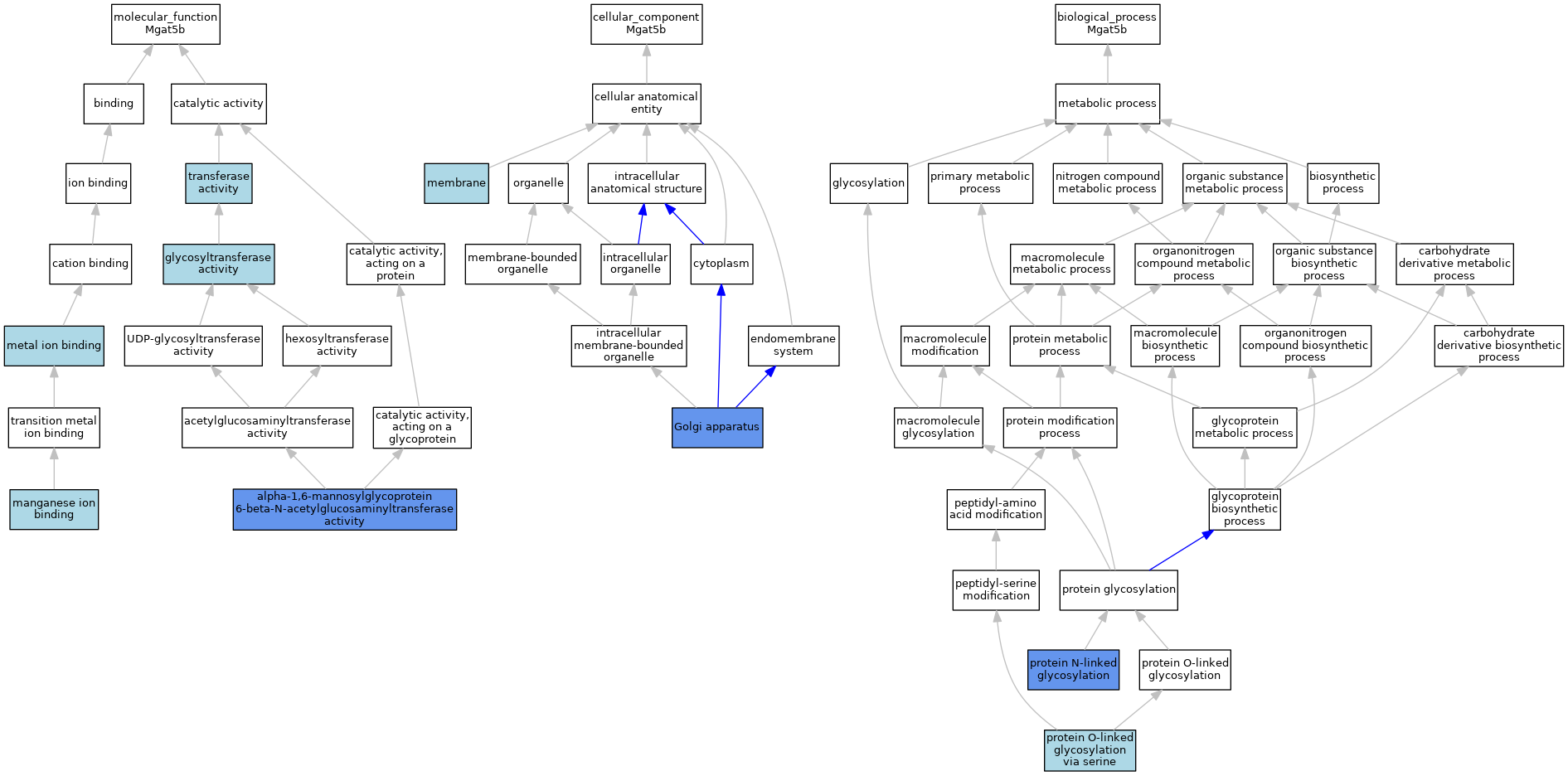
| Molecular Function | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity | IDA | J:112860 | |||||||||
| Molecular Function | GO:0016757 | glycosyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0030145 | manganese ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IDA | J:112860 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0006487 | protein N-linked glycosylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0006487 | protein N-linked glycosylation | IDA | J:112860 | |||||||||
| Biological Process | GO:0018242 | protein O-linked glycosylation via serine | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||