|
Symbol Name ID |
Sppl2c
signal peptide peptidase 2C MGI:3045264 |
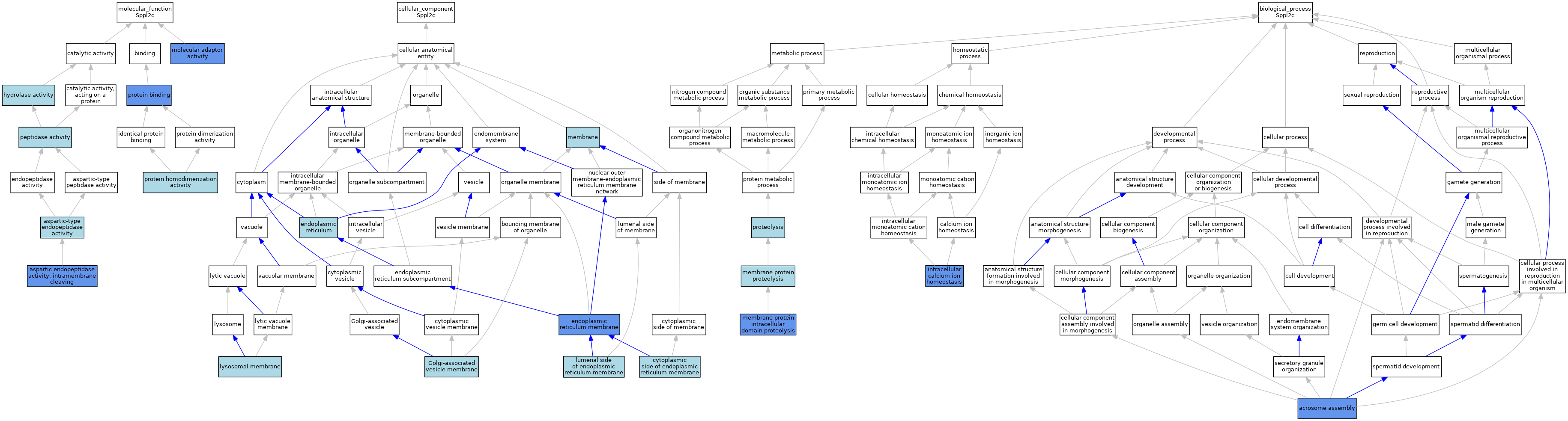
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving | IDA | J:272982 | |||||||||
| Molecular Function | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving | IDA | J:331351 | |||||||||
| Molecular Function | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving | IDA | J:328498 | |||||||||
| Molecular Function | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004190 | aspartic-type endopeptidase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0060090 | molecular adaptor activity | IDA | J:328498 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:328498 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IDA | J:272982 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IDA | J:328498 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IDA | J:331351 | |||||||||
| Cellular Component | GO:0030660 | Golgi-associated vesicle membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0098553 | lumenal side of endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005765 | lysosomal membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0001675 | acrosome assembly | IDA | J:331351 | |||||||||
| Biological Process | GO:0001675 | acrosome assembly | IDA | J:272982 | |||||||||
| Biological Process | GO:0006874 | intracellular calcium ion homeostasis | IDA | J:272982 | |||||||||
| Biological Process | GO:0031293 | membrane protein intracellular domain proteolysis | IDA | J:331351 | |||||||||
| Biological Process | GO:0033619 | membrane protein proteolysis | IBA | J:265628 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||