|
Symbol Name ID |
Elane
elastase, neutrophil expressed MGI:2679229 |
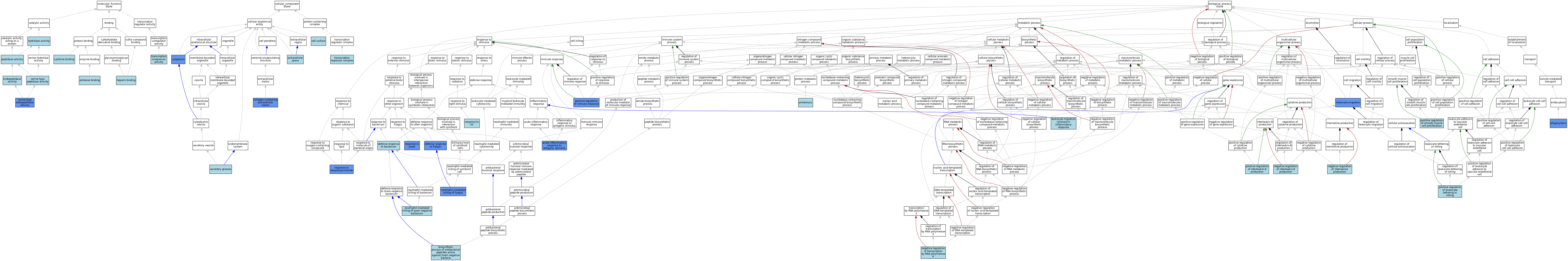
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019955 | cytokine binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004175 | endopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008201 | heparin binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0002020 | protease binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | IDA | J:152360 | |||||||||
| Molecular Function | GO:0008236 | serine-type peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | HDA | J:265604 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:152360 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030141 | secretory granule | ISO | J:73065 | |||||||||
| Cellular Component | GO:0017053 | transcription repressor complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0002438 | acute inflammatory response to antigenic stimulus | IBA | J:265628 | |||||||||
| Biological Process | GO:0002438 | acute inflammatory response to antigenic stimulus | IDA | J:152360 | |||||||||
| Biological Process | GO:0002812 | biosynthetic process of antibacterial peptides active against Gram-negative bacteria | ISO | J:164563 | |||||||||
| Biological Process | GO:0042742 | defense response to bacterium | ISO | J:164563 | |||||||||
| Biological Process | GO:0050832 | defense response to fungus | IMP | J:151389 | |||||||||
| Biological Process | GO:0050832 | defense response to fungus | IGI | J:77597 | |||||||||
| Biological Process | GO:0050832 | defense response to fungus | IMP | J:77597 | |||||||||
| Biological Process | GO:0050900 | leukocyte migration | IMP | J:88722 | |||||||||
| Biological Process | GO:0002523 | leukocyte migration involved in inflammatory response | ISO | J:155856 | |||||||||
| Biological Process | GO:0032682 | negative regulation of chemokine production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032717 | negative regulation of interleukin-8 production | ISO | J:164563 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0070947 | neutrophil-mediated killing of fungus | IMP | J:151389 | |||||||||
| Biological Process | GO:0070945 | neutrophil-mediated killing of gram-negative bacterium | ISO | J:164563 | |||||||||
| Biological Process | GO:0006909 | phagocytosis | IBA | J:265628 | |||||||||
| Biological Process | GO:0006909 | phagocytosis | IMP | J:88722 | |||||||||
| Biological Process | GO:0050778 | positive regulation of immune response | IGI | J:77597 | |||||||||
| Biological Process | GO:0050778 | positive regulation of immune response | IMP | J:77597 | |||||||||
| Biological Process | GO:0032757 | positive regulation of interleukin-8 production | ISO | J:164563 | |||||||||
| Biological Process | GO:1903238 | positive regulation of leukocyte tethering or rolling | ISO | J:155856 | |||||||||
| Biological Process | GO:0048661 | positive regulation of smooth muscle cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IBA | J:265628 | |||||||||
| Biological Process | GO:0006508 | proteolysis | ISO | J:73065 | |||||||||
| Biological Process | GO:0032496 | response to lipopolysaccharide | IGI | J:77597 | |||||||||
| Biological Process | GO:0032496 | response to lipopolysaccharide | IMP | J:77597 | |||||||||
| Biological Process | GO:0009411 | response to UV | ISO | J:164563 | |||||||||
| Biological Process | GO:0001878 | response to yeast | IMP | J:151389 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||