|
Symbol Name ID |
Kiss1
KiSS-1 metastasis-suppressor MGI:2663985 |
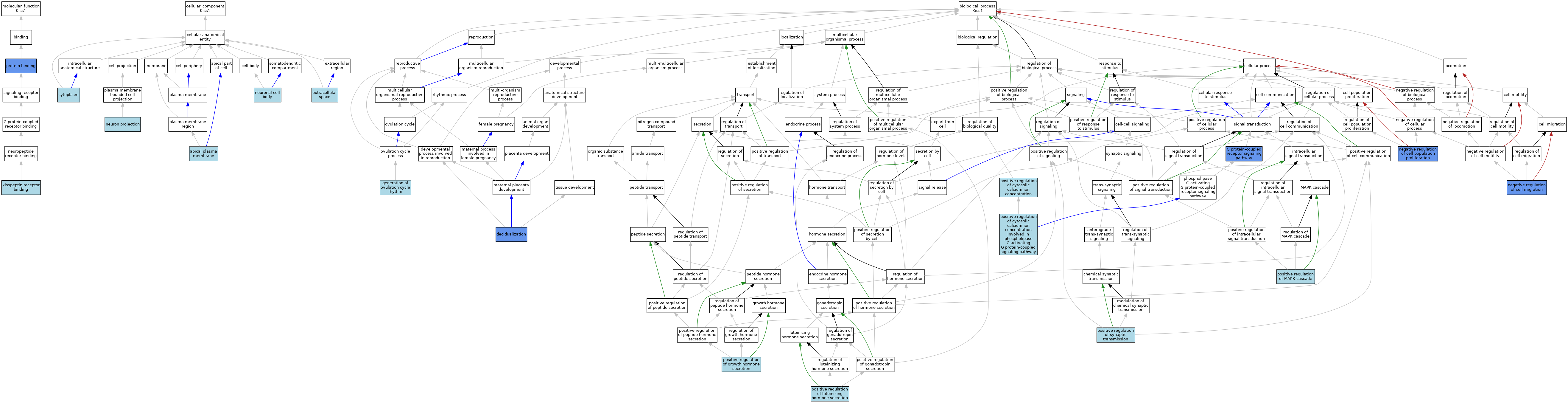
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0031773 | kisspeptin receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0031773 | kisspeptin receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:83519 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:155856 | |||||||||
| Biological Process | GO:0046697 | decidualization | IMP | J:216339 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IDA | J:83519 | |||||||||
| Biological Process | GO:0060112 | generation of ovulation cycle rhythm | ISO | J:155856 | |||||||||
| Biological Process | GO:0030336 | negative regulation of cell migration | IPI | J:83519 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | IPI | J:83519 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | ISO | J:155856 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | IBA | J:265628 | |||||||||
| Biological Process | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0060124 | positive regulation of growth hormone secretion | ISO | J:155856 | |||||||||
| Biological Process | GO:0033686 | positive regulation of luteinizing hormone secretion | ISO | J:155856 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | ISO | J:155856 | |||||||||
| Biological Process | GO:0050806 | positive regulation of synaptic transmission | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||