|
Symbol Name ID |
Ago3
argonaute RISC catalytic subunit 3 MGI:2446634 |
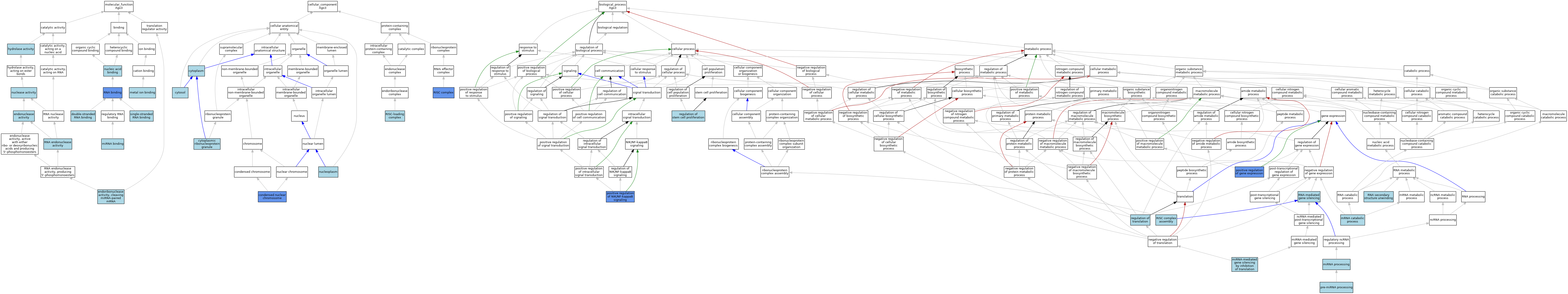
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003725 | double-stranded RNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004519 | endonuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0035198 | miRNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035198 | miRNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004518 | nuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IDA | J:150716 | |||||||||
| Molecular Function | GO:0004521 | RNA endonuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004521 | RNA endonuclease activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003727 | single-stranded RNA binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:187588 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0036464 | cytoplasmic ribonucleoprotein granule | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016442 | RISC complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016442 | RISC complex | IDA | J:150716 | |||||||||
| Cellular Component | GO:0070578 | RISC-loading complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0035196 | miRNA processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006402 | mRNA catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | IMP | J:233964 | |||||||||
| Biological Process | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | IMP | J:233964 | |||||||||
| Biological Process | GO:0031054 | pre-miRNA processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0072091 | regulation of stem cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006417 | regulation of translation | IEA | J:60000 | |||||||||
| Biological Process | GO:0070922 | RISC complex assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0010501 | RNA secondary structure unwinding | ISO | J:164563 | |||||||||
| Biological Process | GO:0031047 | RNA-mediated gene silencing | IEA | J:72247 | |||||||||
| Biological Process | GO:0031047 | RNA-mediated gene silencing | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||