|
Symbol Name ID |
Abra
actin-binding Rho activating protein MGI:2444891 |
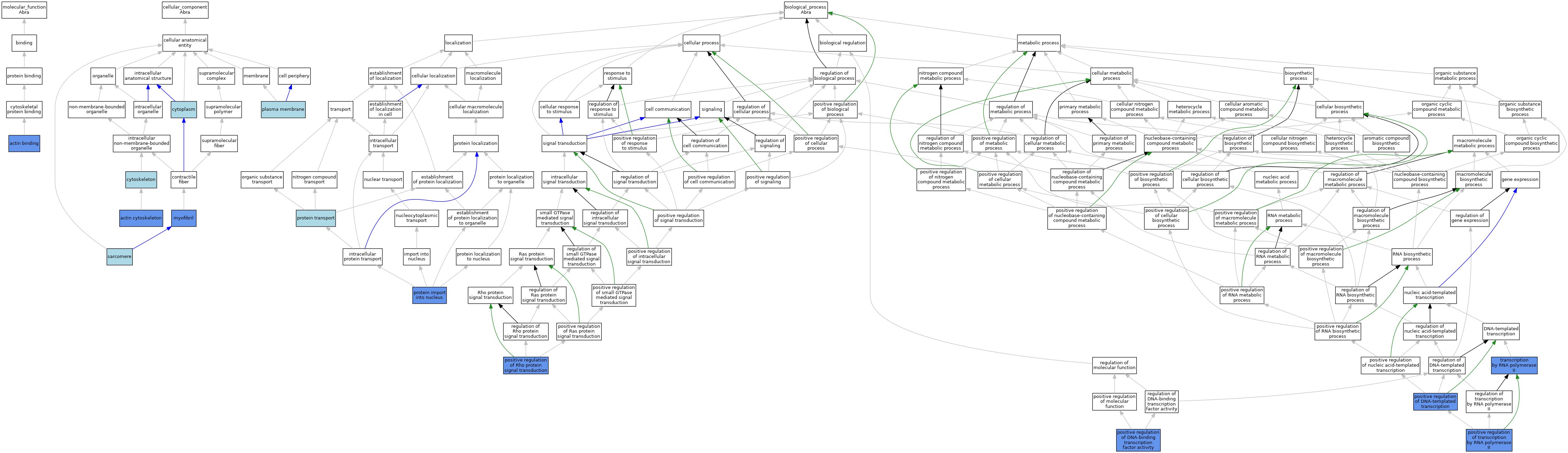
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | IDA | J:116362 | |||||||||
| Cellular Component | GO:0015629 | actin cytoskeleton | IDA | J:116362 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030016 | myofibril | IDA | J:120881 | |||||||||
| Cellular Component | GO:0030016 | myofibril | IDA | J:120881 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030017 | sarcomere | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030017 | sarcomere | IBA | J:265628 | |||||||||
| Biological Process | GO:0051091 | positive regulation of DNA-binding transcription factor activity | IDA | J:116362 | |||||||||
| Biological Process | GO:0051091 | positive regulation of DNA-binding transcription factor activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IDA | J:97653 | |||||||||
| Biological Process | GO:0035025 | positive regulation of Rho protein signal transduction | IDA | J:116362 | |||||||||
| Biological Process | GO:0035025 | positive regulation of Rho protein signal transduction | IBA | J:265628 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IGI | J:120881 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IGI | J:120881 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IDA | J:120881 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IGI | J:120881 | |||||||||
| Biological Process | GO:0006606 | protein import into nucleus | IDA | J:97653 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:120881 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IDA | J:120881 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:120881 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IDA | J:120881 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:120881 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:120881 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:120881 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:120881 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||