|
Symbol Name ID |
Mapkap1
mitogen-activated protein kinase associated protein 1 MGI:2444554 |
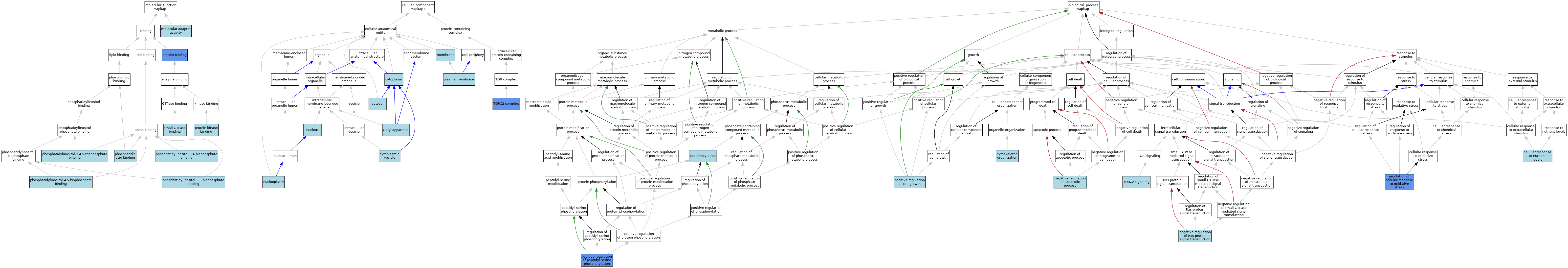
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0060090 | molecular adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0070300 | phosphatidic acid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:167176 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:172256 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:172256 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031267 | small GTPase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031932 | TORC2 complex | IDA | J:125618 | |||||||||
| Cellular Component | GO:0031932 | TORC2 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031932 | TORC2 complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0031669 | cellular response to nutrient levels | NAS | J:319697 | |||||||||
| Biological Process | GO:0007010 | cytoskeleton organization | NAS | J:320275 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | NAS | J:319697 | |||||||||
| Biological Process | GO:0046580 | negative regulation of Ras protein signal transduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0030307 | positive regulation of cell growth | NAS | J:319697 | |||||||||
| Biological Process | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | IMP | J:125618 | |||||||||
| Biological Process | GO:1900407 | regulation of cellular response to oxidative stress | IMP | J:125618 | |||||||||
| Biological Process | GO:0038203 | TORC2 signaling | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||