|
Symbol Name ID |
Gimap5
GTPase, IMAP family member 5 MGI:2442232 |
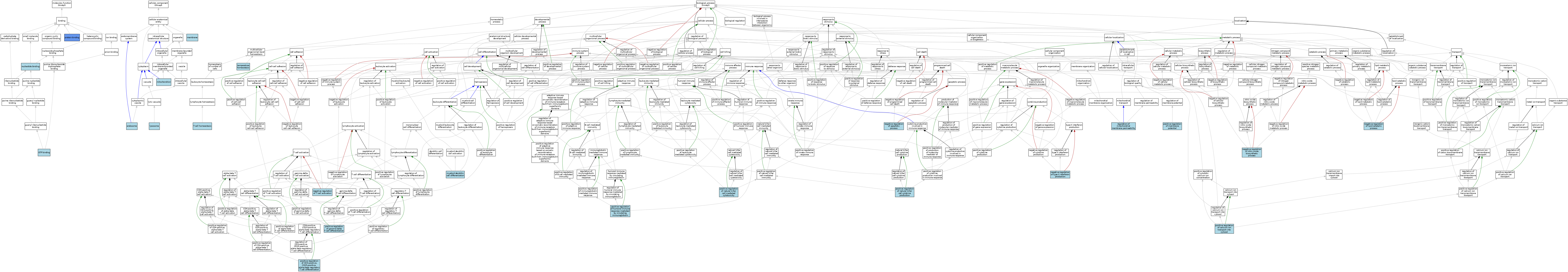
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005525 | GTP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:107746 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Biological Process | GO:0043011 | myeloid dendritic cell differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0050995 | negative regulation of lipid catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0045019 | negative regulation of nitric oxide biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0050868 | negative regulation of T cell activation | ISO | J:155856 | |||||||||
| Biological Process | GO:0032689 | negative regulation of type II interferon production | ISO | J:155856 | |||||||||
| Biological Process | GO:0010524 | positive regulation of calcium ion transport into cytosol | ISO | J:155856 | |||||||||
| Biological Process | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0045588 | positive regulation of gamma-delta T cell differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin | ISO | J:155856 | |||||||||
| Biological Process | GO:0045838 | positive regulation of membrane potential | ISO | J:155856 | |||||||||
| Biological Process | GO:0002729 | positive regulation of natural killer cell cytokine production | ISO | J:155856 | |||||||||
| Biological Process | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | ISO | J:155856 | |||||||||
| Biological Process | GO:0046902 | regulation of mitochondrial membrane permeability | ISO | J:155856 | |||||||||
| Biological Process | GO:0043029 | T cell homeostasis | ISO | J:155856 | |||||||||
| Biological Process | GO:0001659 | temperature homeostasis | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||