|
Symbol Name ID |
Wwc1
WW, C2 and coiled-coil domain containing 1 MGI:2388637 |
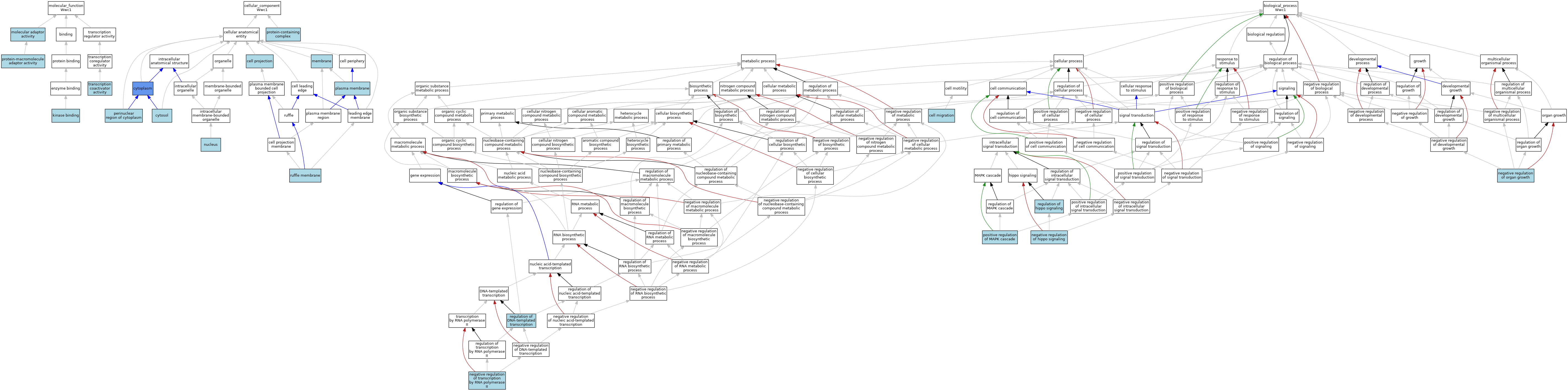
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019900 | kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019900 | kinase binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0060090 | molecular adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0060090 | molecular adaptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0030674 | protein-macromolecule adaptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:133423 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0032587 | ruffle membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0016477 | cell migration | ISO | J:155856 | |||||||||
| Biological Process | GO:0016477 | cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0016477 | cell migration | IBA | J:265628 | |||||||||
| Biological Process | GO:0035331 | negative regulation of hippo signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0046621 | negative regulation of organ growth | ISO | J:164563 | |||||||||
| Biological Process | GO:0046621 | negative regulation of organ growth | IBA | J:265628 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IBA | J:265628 | |||||||||
| Biological Process | GO:0035330 | regulation of hippo signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0035330 | regulation of hippo signaling | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||