|
Symbol Name ID |
Plekhn1
pleckstrin homology domain containing, family N member 1 MGI:2387630 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
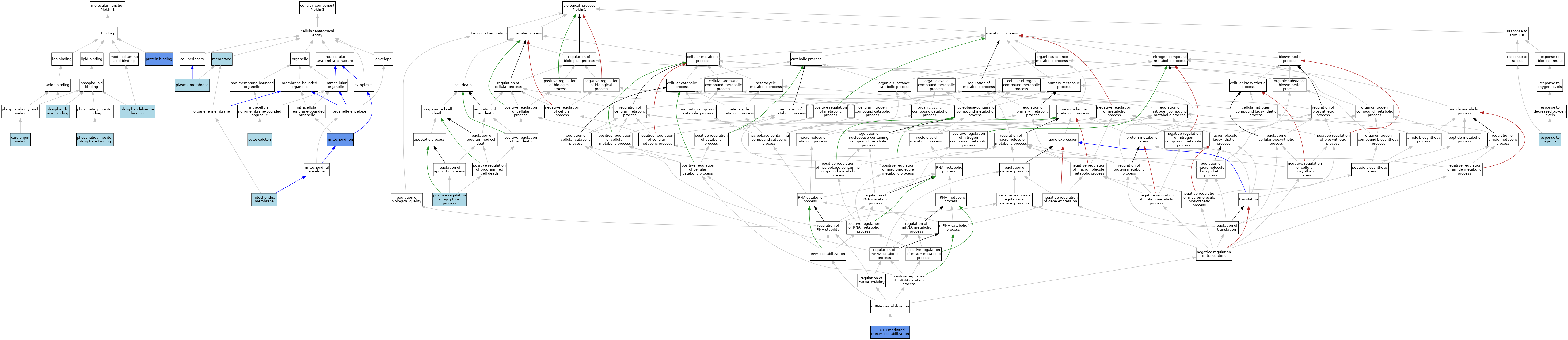
| Molecular Function | GO:1901612 | cardiolipin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:1901612 | cardiolipin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0070300 | phosphatidic acid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0070300 | phosphatidic acid binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:1901981 | phosphatidylinositol phosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:1901981 | phosphatidylinositol phosphate binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0001786 | phosphatidylserine binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001786 | phosphatidylserine binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114108 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031966 | mitochondrial membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031966 | mitochondrial membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:251603 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0061158 | 3'-UTR-mediated mRNA destabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0061158 | 3'-UTR-mediated mRNA destabilization | IDA | J:251603 | |||||||||
| Biological Process | GO:0061158 | 3'-UTR-mediated mRNA destabilization | IBA | J:265628 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0001666 | response to hypoxia | ISO | J:164563 | |||||||||
| Biological Process | GO:0001666 | response to hypoxia | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||