|
Symbol Name ID |
Tardbp
TAR DNA binding protein MGI:2387629 |
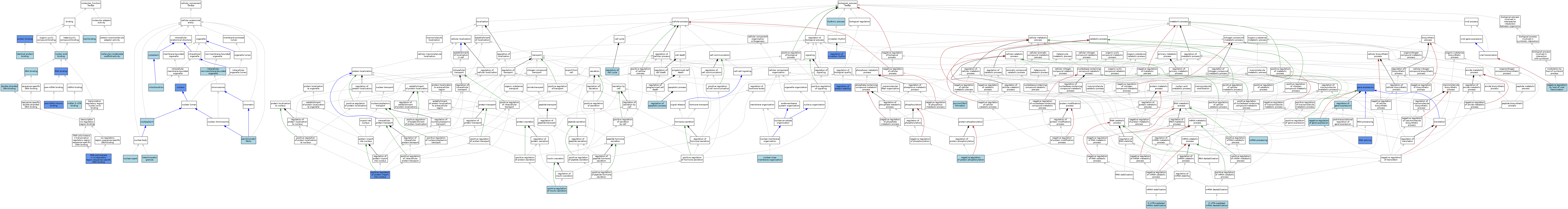
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003690 | double-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0140693 | molecular condensate scaffold activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003730 | mRNA 3'-UTR binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0097157 | pre-mRNA intronic binding | IDA | J:240782 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:292536 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:250353 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:190257 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:193640 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IDA | J:240782 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IDA | J:186828 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0035061 | interchromatin granule | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043232 | intracellular non-membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:250353 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005726 | perichromatin fibrils | ISO | J:155856 | |||||||||
| Biological Process | GO:0061158 | 3'-UTR-mediated mRNA destabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0070935 | 3'-UTR-mediated mRNA stabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:1990000 | amyloid fibril formation | ISO | J:164563 | |||||||||
| Biological Process | GO:0010467 | gene expression | IMP | J:240782 | |||||||||
| Biological Process | GO:0006397 | mRNA processing | IEA | J:60000 | |||||||||
| Biological Process | GO:0043922 | negative regulation by host of viral transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:0001933 | negative regulation of protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0071765 | nuclear inner membrane organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0032024 | positive regulation of insulin secretion | ISO | J:155856 | |||||||||
| Biological Process | GO:0042307 | positive regulation of protein import into nucleus | IDA | J:250353 | |||||||||
| Biological Process | GO:0042981 | regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0051726 | regulation of cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0042752 | regulation of circadian rhythm | IMP | J:283196 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IBA | J:265628 | |||||||||
| Biological Process | GO:0031647 | regulation of protein stability | ISO | J:164563 | |||||||||
| Biological Process | GO:0031647 | regulation of protein stability | IMP | J:283196 | |||||||||
| Biological Process | GO:0048511 | rhythmic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0008380 | RNA splicing | ISO | J:164563 | |||||||||
| Biological Process | GO:0008380 | RNA splicing | IMP | J:240782 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||