|
Symbol Name ID |
Spg7
SPG7, paraplegin matrix AAA peptidase subunit MGI:2385906 |
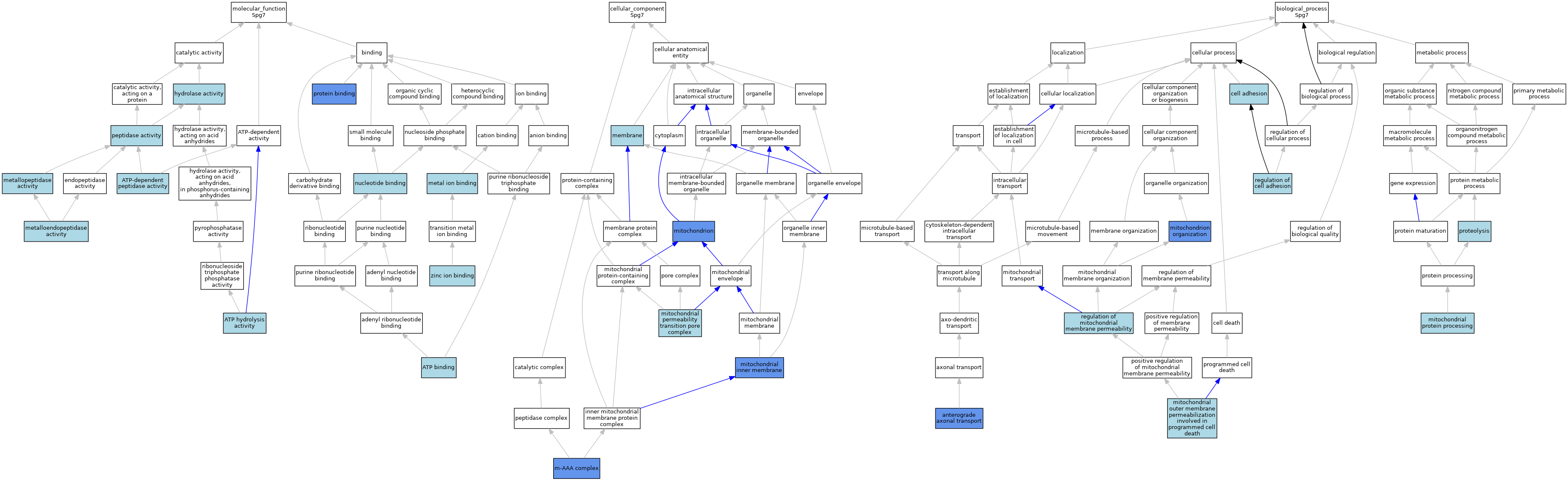
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004176 | ATP-dependent peptidase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008237 | metallopeptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:164484 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005745 | m-AAA complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005745 | m-AAA complex | IDA | J:164484 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005743 | mitochondrial inner membrane | IDA | J:164484 | |||||||||
| Cellular Component | GO:0005757 | mitochondrial permeability transition pore complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:164484 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:87616 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Biological Process | GO:0008089 | anterograde axonal transport | IMP | J:87616 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | TAS | J:45282 | |||||||||
| Biological Process | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | ISO | J:164563 | |||||||||
| Biological Process | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | ISO | J:155856 | |||||||||
| Biological Process | GO:0034982 | mitochondrial protein processing | IBA | J:265628 | |||||||||
| Biological Process | GO:0007005 | mitochondrion organization | IMP | J:87616 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:72247 | |||||||||
| Biological Process | GO:0030155 | regulation of cell adhesion | TAS | J:45282 | |||||||||
| Biological Process | GO:0046902 | regulation of mitochondrial membrane permeability | ISO | J:155856 | |||||||||
| Biological Process | GO:0046902 | regulation of mitochondrial membrane permeability | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||