|
Symbol Name ID |
Dhrs7b
dehydrogenase/reductase 7B MGI:2384931 |
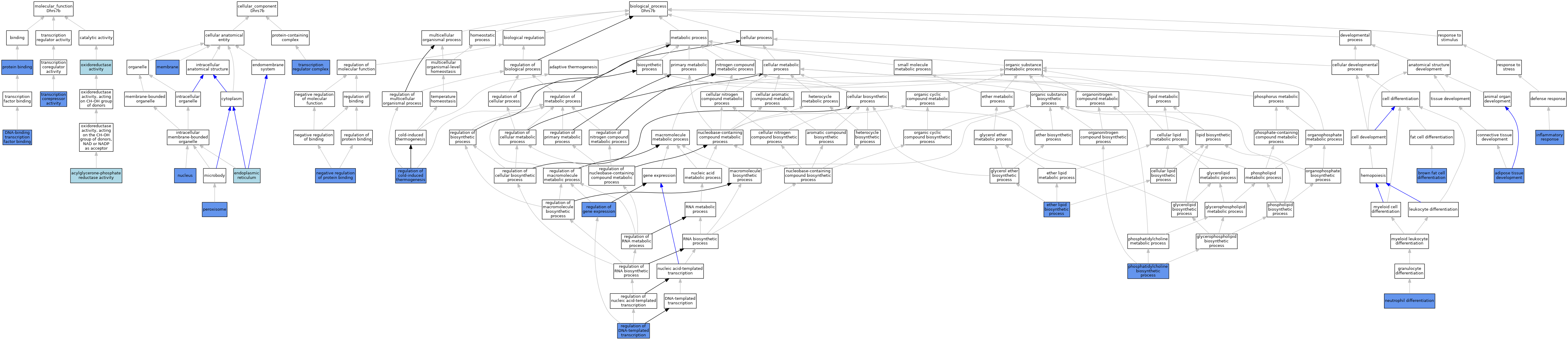
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0000140 | acylglycerone-phosphate reductase activity | TAS | J:219976 | |||||||||
| Molecular Function | GO:0140297 | DNA-binding transcription factor binding | IPI | J:251809 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:251809 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:251809 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:251809 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:251809 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IMP | J:251809 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IDA | J:251809 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:251809 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:251809 | |||||||||
| Cellular Component | GO:0005777 | peroxisome | IDA | J:251809 | |||||||||
| Cellular Component | GO:0005667 | transcription regulator complex | IPI | J:251809 | |||||||||
| Biological Process | GO:0060612 | adipose tissue development | IMP | J:251809 | |||||||||
| Biological Process | GO:0050873 | brown fat cell differentiation | IDA | J:251809 | |||||||||
| Biological Process | GO:0008611 | ether lipid biosynthetic process | IMP | J:219976 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IMP | J:219976 | |||||||||
| Biological Process | GO:0032091 | negative regulation of protein binding | IDA | J:251809 | |||||||||
| Biological Process | GO:0030223 | neutrophil differentiation | IMP | J:219976 | |||||||||
| Biological Process | GO:0006656 | phosphatidylcholine biosynthetic process | IMP | J:251809 | |||||||||
| Biological Process | GO:0120161 | regulation of cold-induced thermogenesis | IMP | J:251809 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IDA | J:251809 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IMP | J:251809 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IMP | J:251809 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IMP | J:251809 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||