|
Symbol Name ID |
Adgrf5
adhesion G protein-coupled receptor F5 MGI:2182928 |
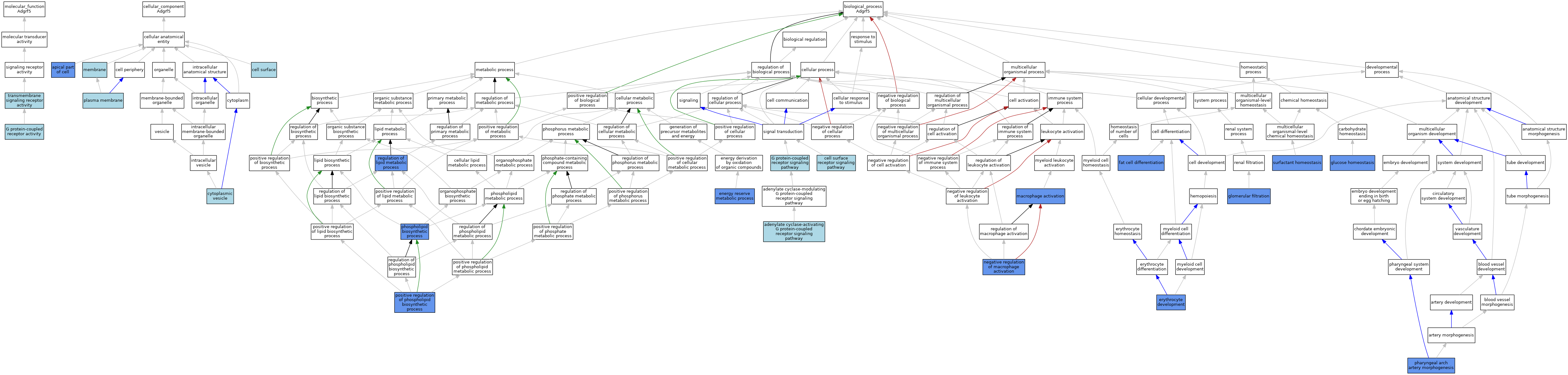
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004888 | transmembrane signaling receptor activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | IDA | J:221769 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | IDA | J:221769 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | IDA | J:221769 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:73065 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | ISO | J:73065 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007166 | cell surface receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0006112 | energy reserve metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006112 | energy reserve metabolic process | IMP | J:190131 | |||||||||
| Biological Process | GO:0048821 | erythrocyte development | IGI | J:245698 | |||||||||
| Biological Process | GO:0045444 | fat cell differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0045444 | fat cell differentiation | IMP | J:190131 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0003094 | glomerular filtration | IGI | J:245698 | |||||||||
| Biological Process | GO:0042593 | glucose homeostasis | IMP | J:190131 | |||||||||
| Biological Process | GO:0042116 | macrophage activation | IMP | J:222167 | |||||||||
| Biological Process | GO:0043031 | negative regulation of macrophage activation | IMP | J:222167 | |||||||||
| Biological Process | GO:0061626 | pharyngeal arch artery morphogenesis | IGI | J:245698 | |||||||||
| Biological Process | GO:0008654 | phospholipid biosynthetic process | IMP | J:221769 | |||||||||
| Biological Process | GO:0071073 | positive regulation of phospholipid biosynthetic process | IMP | J:221769 | |||||||||
| Biological Process | GO:0019216 | regulation of lipid metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0019216 | regulation of lipid metabolic process | IMP | J:190131 | |||||||||
| Biological Process | GO:0043129 | surfactant homeostasis | IMP | J:245698 | |||||||||
| Biological Process | GO:0043129 | surfactant homeostasis | IMP | J:221769 | |||||||||
| Biological Process | GO:0043129 | surfactant homeostasis | IMP | J:196943 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||