|
Symbol Name ID |
Trrap
transformation/transcription domain-associated protein MGI:2153272 |
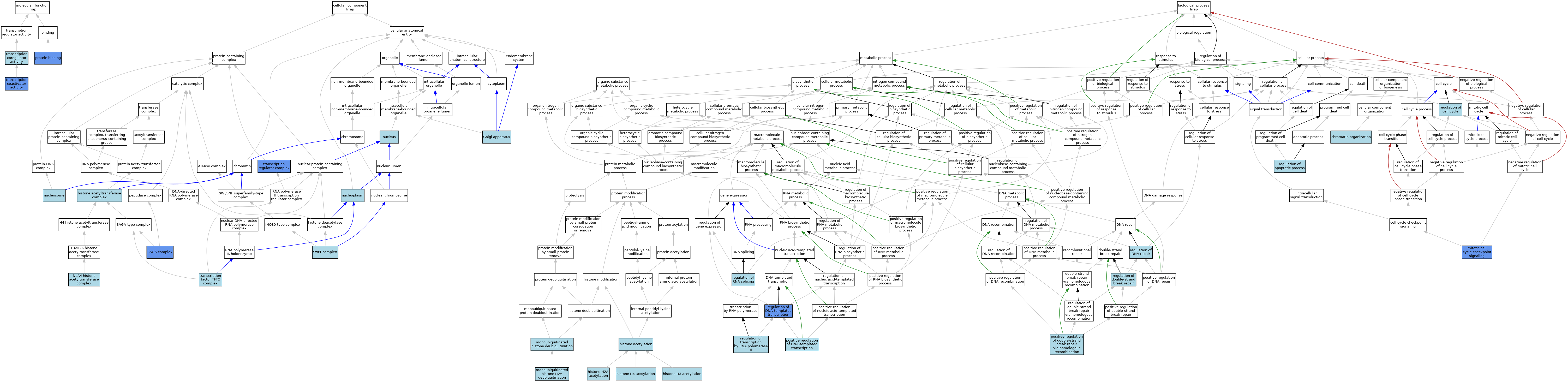
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | IPI | J:162104 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:107098 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:162104 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:107098 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:168649 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | IDA | J:80691 | |||||||||
| Molecular Function | GO:0003712 | transcription coregulator activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000123 | histone acetyltransferase complex | TAS | J:71950 | |||||||||
| Cellular Component | GO:0035267 | NuA4 histone acetyltransferase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000786 | nucleosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000124 | SAGA complex | NAS | J:320037 | |||||||||
| Cellular Component | GO:0000124 | SAGA complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000124 | SAGA complex | IDA | J:245730 | |||||||||
| Cellular Component | GO:0000812 | Swr1 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0033276 | transcription factor TFTC complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0033276 | transcription factor TFTC complex | NAS | J:320038 | |||||||||
| Cellular Component | GO:0005667 | transcription regulator complex | IDA | J:80691 | |||||||||
| Biological Process | GO:0006325 | chromatin organization | IEA | J:60000 | |||||||||
| Biological Process | GO:0016573 | histone acetylation | NAS | J:320277 | |||||||||
| Biological Process | GO:0016573 | histone acetylation | NAS | J:320265 | |||||||||
| Biological Process | GO:0043968 | histone H2A acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043966 | histone H3 acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043966 | histone H3 acetylation | NAS | J:320036 | |||||||||
| Biological Process | GO:0043966 | histone H3 acetylation | NAS | J:320278 | |||||||||
| Biological Process | GO:0043967 | histone H4 acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0007093 | mitotic cell cycle checkpoint signaling | IMP | J:71950 | |||||||||
| Biological Process | GO:0035521 | monoubiquitinated histone deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0035522 | monoubiquitinated histone H2A deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | NAS | J:319957 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | NAS | J:320111 | |||||||||
| Biological Process | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0042981 | regulation of apoptotic process | NAS | J:320265 | |||||||||
| Biological Process | GO:0051726 | regulation of cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0006282 | regulation of DNA repair | NAS | J:320036 | |||||||||
| Biological Process | GO:0006282 | regulation of DNA repair | NAS | J:320001 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | NAS | J:320276 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IDA | J:80691 | |||||||||
| Biological Process | GO:2000779 | regulation of double-strand break repair | NAS | J:320265 | |||||||||
| Biological Process | GO:0043484 | regulation of RNA splicing | NAS | J:320036 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||