|
Symbol Name ID |
Afap1l2
actin filament associated protein 1-like 2 MGI:2147658 |
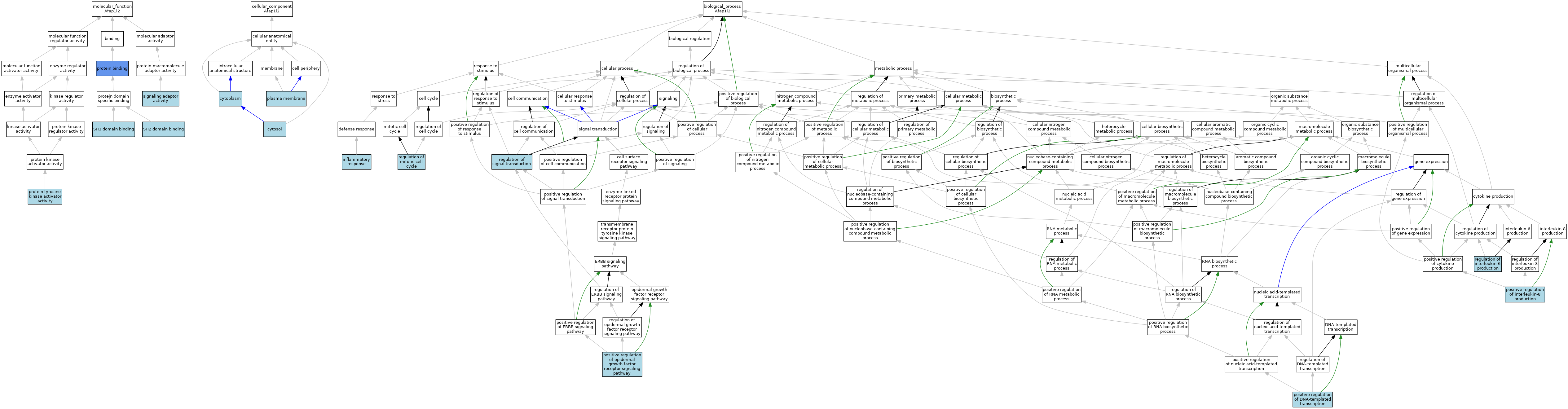
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | IPI | J:273694 | |||||||||
| Molecular Function | GO:0030296 | protein tyrosine kinase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042169 | SH2 domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042169 | SH2 domain binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0035591 | signaling adaptor activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IBA | J:265628 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IBA | J:265628 | |||||||||
| Biological Process | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0032757 | positive regulation of interleukin-8 production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032757 | positive regulation of interleukin-8 production | IBA | J:265628 | |||||||||
| Biological Process | GO:0032675 | regulation of interleukin-6 production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032675 | regulation of interleukin-6 production | IBA | J:265628 | |||||||||
| Biological Process | GO:0007346 | regulation of mitotic cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0007346 | regulation of mitotic cell cycle | IBA | J:265628 | |||||||||
| Biological Process | GO:0009966 | regulation of signal transduction | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||