|
Symbol Name ID |
Adgrb1
adhesion G protein-coupled receptor B1 MGI:1933736 |
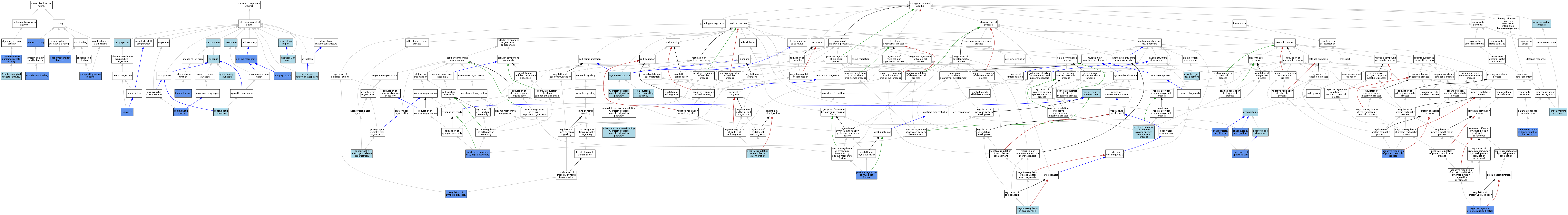
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0001530 | lipopolysaccharide binding | IDA | J:169123 | |||||||||
| Molecular Function | GO:0030165 | PDZ domain binding | IPI | J:202713 | |||||||||
| Molecular Function | GO:0001786 | phosphatidylserine binding | IDA | J:127717 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:127717 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:222100 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:127717 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:68103 | |||||||||
| Molecular Function | GO:0004888 | transmembrane signaling receptor activity | IDA | J:127717 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IDA | J:243397 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IDA | J:243397 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0001891 | phagocytic cup | IDA | J:127717 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:127717 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:243397 | |||||||||
| Cellular Component | GO:0014069 | postsynaptic density | IDA | J:196966 | |||||||||
| Cellular Component | GO:0014069 | postsynaptic density | IDA | J:202713 | |||||||||
| Cellular Component | GO:0014069 | postsynaptic density | IBA | J:265628 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IEA | J:60000 | |||||||||
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0043277 | apoptotic cell clearance | ISO | J:164563 | |||||||||
| Biological Process | GO:0007166 | cell surface receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0050829 | defense response to Gram-negative bacterium | IMP | J:169123 | |||||||||
| Biological Process | GO:0043652 | engulfment of apoptotic cell | IDA | J:127717 | |||||||||
| Biological Process | GO:0043652 | engulfment of apoptotic cell | ISO | J:164563 | |||||||||
| Biological Process | GO:0043652 | engulfment of apoptotic cell | IBA | J:265628 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:60000 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0007517 | muscle organ development | IEA | J:60000 | |||||||||
| Biological Process | GO:0016525 | negative regulation of angiogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0016525 | negative regulation of angiogenesis | ISO | J:68103 | |||||||||
| Biological Process | GO:0010596 | negative regulation of endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0042177 | negative regulation of protein catabolic process | IMP | J:222100 | |||||||||
| Biological Process | GO:0031397 | negative regulation of protein ubiquitination | IMP | J:222100 | |||||||||
| Biological Process | GO:0007399 | nervous system development | IEA | J:60000 | |||||||||
| Biological Process | GO:0006909 | phagocytosis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006911 | phagocytosis, engulfment | IMP | J:243461 | |||||||||
| Biological Process | GO:0006910 | phagocytosis, recognition | IMP | J:169123 | |||||||||
| Biological Process | GO:1901741 | positive regulation of myoblast fusion | IMP | J:198690 | |||||||||
| Biological Process | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0051965 | positive regulation of synapse assembly | IDA | J:211830 | |||||||||
| Biological Process | GO:0098974 | postsynaptic actin cytoskeleton organization | ISO | J:155856 | |||||||||
| Biological Process | GO:0048167 | regulation of synaptic plasticity | IMP | J:222100 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||