|
Symbol Name ID |
Cubn
cubilin MGI:1931256 |
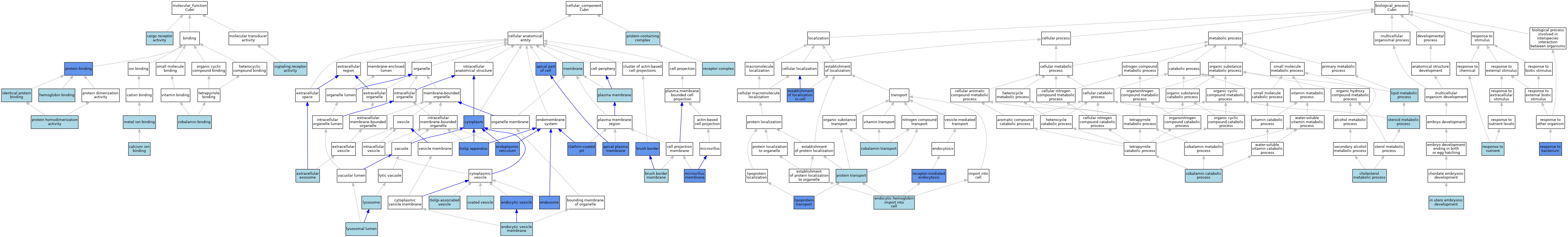
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0038024 | cargo receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0038024 | cargo receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0038024 | cargo receptor activity | ISO | J:311657 | |||||||||
| Molecular Function | GO:0031419 | cobalamin binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0030492 | hemoglobin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:76353 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0038023 | signaling receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0038023 | signaling receptor activity | TAS | J:67664 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | IDA | J:98337 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | IDA | J:76353 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | IDA | J:98256 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | NAS | J:311657 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | IDA | J:98256 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | IDA | J:98256 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | IDA | J:98256 | |||||||||
| Cellular Component | GO:0005903 | brush border | IDA | J:84433 | |||||||||
| Cellular Component | GO:0031526 | brush border membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005905 | clathrin-coated pit | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005905 | clathrin-coated pit | IDA | J:98256 | |||||||||
| Cellular Component | GO:0030135 | coated vesicle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:76353 | |||||||||
| Cellular Component | GO:0030139 | endocytic vesicle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030139 | endocytic vesicle | IDA | J:98256 | |||||||||
| Cellular Component | GO:0030666 | endocytic vesicle membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:98256 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:98256 | |||||||||
| Cellular Component | GO:0005768 | endosome | IDA | J:67664 | |||||||||
| Cellular Component | GO:0005768 | endosome | IDA | J:84433 | |||||||||
| Cellular Component | GO:0070062 | extracellular exosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IDA | J:98256 | |||||||||
| Cellular Component | GO:0005798 | Golgi-associated vesicle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043202 | lysosomal lumen | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0031528 | microvillus membrane | IDA | J:319880 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0008203 | cholesterol metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0042366 | cobalamin catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0015889 | cobalamin transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0015889 | cobalamin transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0015889 | cobalamin transport | ISO | J:311657 | |||||||||
| Biological Process | GO:0020028 | endocytic hemoglobin import into cell | ISO | J:155856 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IDA | J:98256 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IMP | J:98256 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | ISO | J:155856 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0042953 | lipoprotein transport | IDA | J:98256 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0006898 | receptor-mediated endocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0006898 | receptor-mediated endocytosis | IMP | J:98256 | |||||||||
| Biological Process | GO:0009617 | response to bacterium | IEP | J:190322 | |||||||||
| Biological Process | GO:0007584 | response to nutrient | ISO | J:155856 | |||||||||
| Biological Process | GO:0008202 | steroid metabolic process | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||