|
Symbol Name ID |
Dnajb2
DnaJ heat shock protein family (Hsp40) member B2 MGI:1928739 |
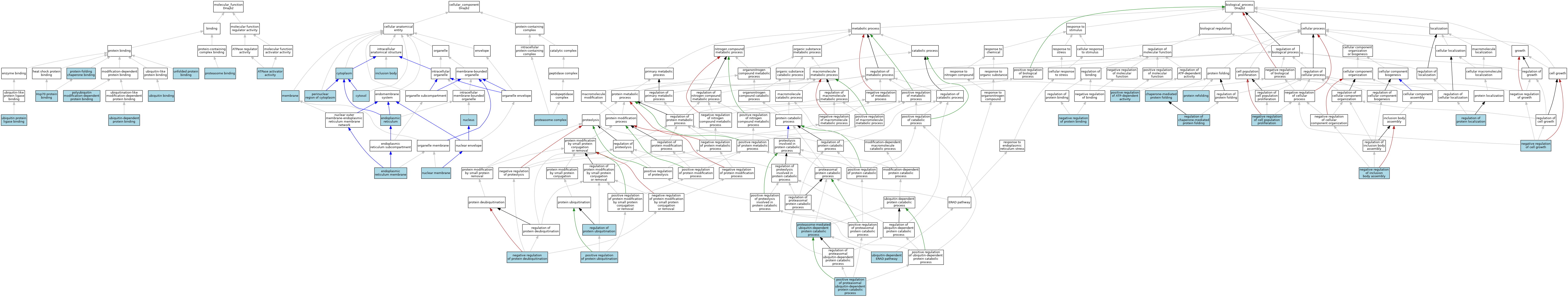
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0001671 | ATPase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001671 | ATPase activator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0030544 | Hsp70 protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030544 | Hsp70 protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030544 | Hsp70 protein binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031593 | polyubiquitin modification-dependent protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0070628 | proteasome binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043130 | ubiquitin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0140036 | ubiquitin-dependent protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051082 | unfolded protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051082 | unfolded protein binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016234 | inclusion body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000502 | proteasome complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0061077 | chaperone-mediated protein folding | ISO | J:164563 | |||||||||
| Biological Process | GO:0061077 | chaperone-mediated protein folding | IBA | J:265628 | |||||||||
| Biological Process | GO:0030308 | negative regulation of cell growth | ISO | J:164563 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0090084 | negative regulation of inclusion body assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0090084 | negative regulation of inclusion body assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0032091 | negative regulation of protein binding | ISO | J:164563 | |||||||||
| Biological Process | GO:0090086 | negative regulation of protein deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0032781 | positive regulation of ATP-dependent activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0031398 | positive regulation of protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0042026 | protein refolding | ISO | J:164563 | |||||||||
| Biological Process | GO:0042026 | protein refolding | IBA | J:265628 | |||||||||
| Biological Process | GO:1903644 | regulation of chaperone-mediated protein folding | ISO | J:164563 | |||||||||
| Biological Process | GO:0032880 | regulation of protein localization | ISO | J:164563 | |||||||||
| Biological Process | GO:0031396 | regulation of protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0030433 | ubiquitin-dependent ERAD pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0030433 | ubiquitin-dependent ERAD pathway | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||