|
Symbol Name ID |
Arl6
ADP-ribosylation factor-like 6 MGI:1927136 |
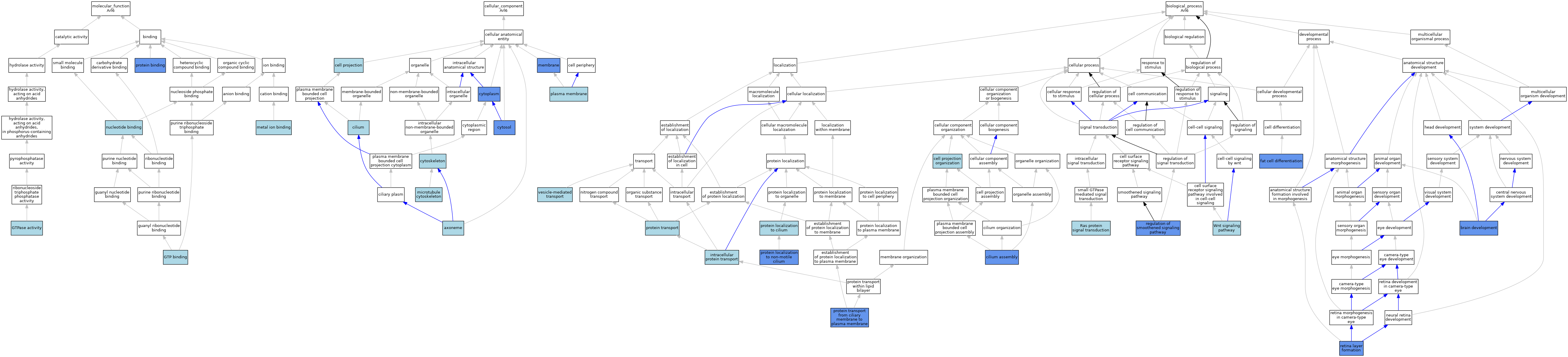
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005525 | GTP binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | ISS | J:62458 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62458 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62458 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62458 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62458 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62458 | |||||||||
| Cellular Component | GO:0005930 | axoneme | IBA | J:265628 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005929 | cilium | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:62458 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:180524 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:62458 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:180524 | |||||||||
| Cellular Component | GO:0015630 | microtubule cytoskeleton | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:180524 | |||||||||
| Biological Process | GO:0030030 | cell projection organization | IEA | J:60000 | |||||||||
| Biological Process | GO:0060271 | cilium assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0060271 | cilium assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0060271 | cilium assembly | IMP | J:180524 | |||||||||
| Biological Process | GO:0045444 | fat cell differentiation | IEP | J:135549 | |||||||||
| Biological Process | GO:0006886 | intracellular protein transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0061512 | protein localization to cilium | IBA | J:265628 | |||||||||
| Biological Process | GO:0061512 | protein localization to cilium | ISO | J:73065 | |||||||||
| Biological Process | GO:0097499 | protein localization to non-motile cilium | IDA | J:214729 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:1903445 | protein transport from ciliary membrane to plasma membrane | IMP | J:180524 | |||||||||
| Biological Process | GO:0007265 | Ras protein signal transduction | ISS | J:62458 | |||||||||
| Biological Process | GO:0008589 | regulation of smoothened signaling pathway | IMP | J:180524 | |||||||||
| Biological Process | GO:0010842 | retina layer formation | IMP | J:180524 | |||||||||
| Biological Process | GO:0010842 | retina layer formation | IMP | J:159222 | |||||||||
| Biological Process | GO:0016192 | vesicle-mediated transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0016055 | Wnt signaling pathway | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||