|
Symbol Name ID |
Tnip1
TNFAIP3 interacting protein 1 MGI:1926194 |
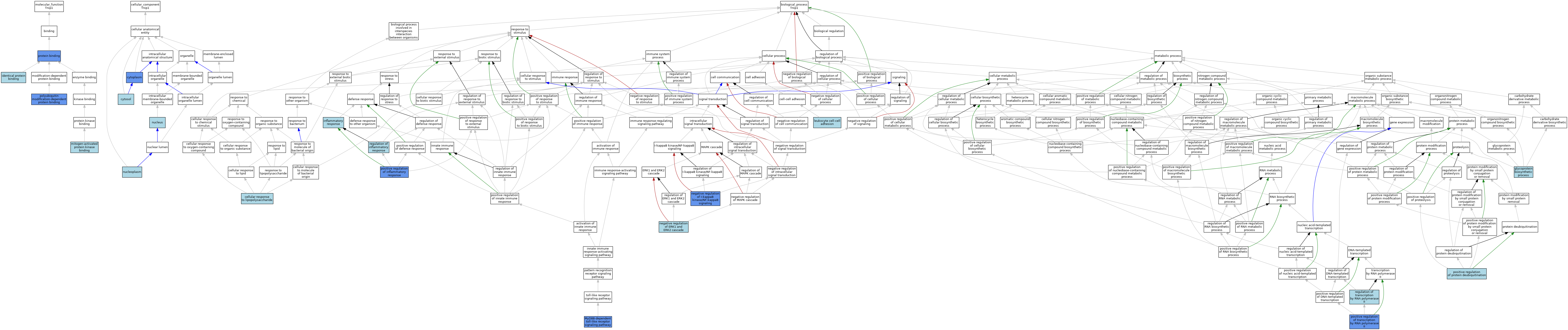
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051019 | mitogen-activated protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051019 | mitogen-activated protein kinase binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031593 | polyubiquitin modification-dependent protein binding | IDA | J:183962 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:180054 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:145539 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:186074 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:64942 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:64942 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Biological Process | GO:0071222 | cellular response to lipopolysaccharide | IBA | J:265628 | |||||||||
| Biological Process | GO:0009101 | glycoprotein biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IEA | J:60000 | |||||||||
| Biological Process | GO:0007159 | leukocyte cell-cell adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | IDA | J:180054 | |||||||||
| Biological Process | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | IBA | J:265628 | |||||||||
| Biological Process | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | IDA | J:183962 | |||||||||
| Biological Process | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0050729 | positive regulation of inflammatory response | IMP | J:180054 | |||||||||
| Biological Process | GO:1903003 | positive regulation of protein deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:180054 | |||||||||
| Biological Process | GO:0050727 | regulation of inflammatory response | IEA | J:72247 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||