|
Symbol Name ID |
Ak7
adenylate kinase 7 MGI:1926051 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
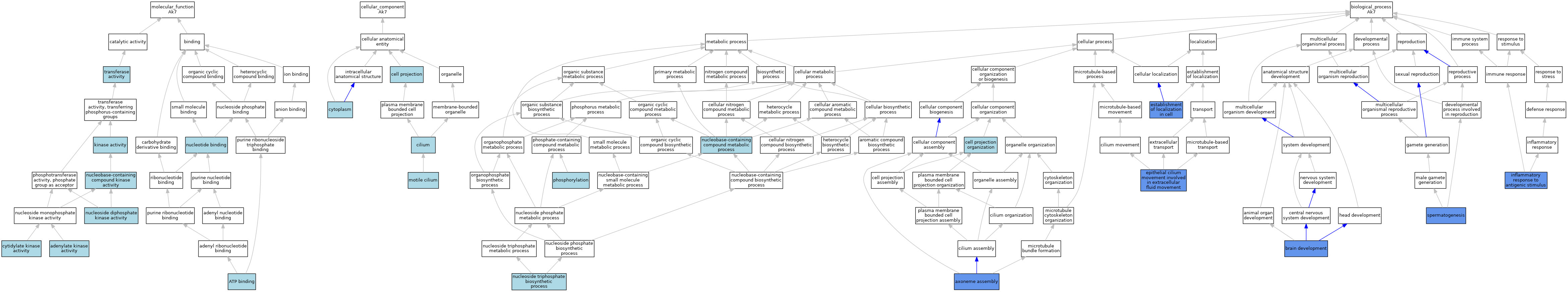
| Molecular Function | GO:0004017 | adenylate kinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004017 | adenylate kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004127 | cytidylate kinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004127 | cytidylate kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0019205 | nucleobase-containing compound kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004550 | nucleoside diphosphate kinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004550 | nucleoside diphosphate kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005929 | cilium | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031514 | motile cilium | IEA | J:60000 | |||||||||
| Biological Process | GO:0035082 | axoneme assembly | IMP | J:186062 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:186062 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:186062 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:185566 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:186062 | |||||||||
| Biological Process | GO:0030030 | cell projection organization | IEA | J:60000 | |||||||||
| Biological Process | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | IMP | J:185566 | |||||||||
| Biological Process | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | IMP | J:185566 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IMP | J:185566 | |||||||||
| Biological Process | GO:0002437 | inflammatory response to antigenic stimulus | IMP | J:186062 | |||||||||
| Biological Process | GO:0006139 | nucleobase-containing compound metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0009142 | nucleoside triphosphate biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0009142 | nucleoside triphosphate biosynthetic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:186062 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:186062 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||