|
Symbol Name ID |
Rnase6
ribonuclease, RNase A family, 6 MGI:1925666 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
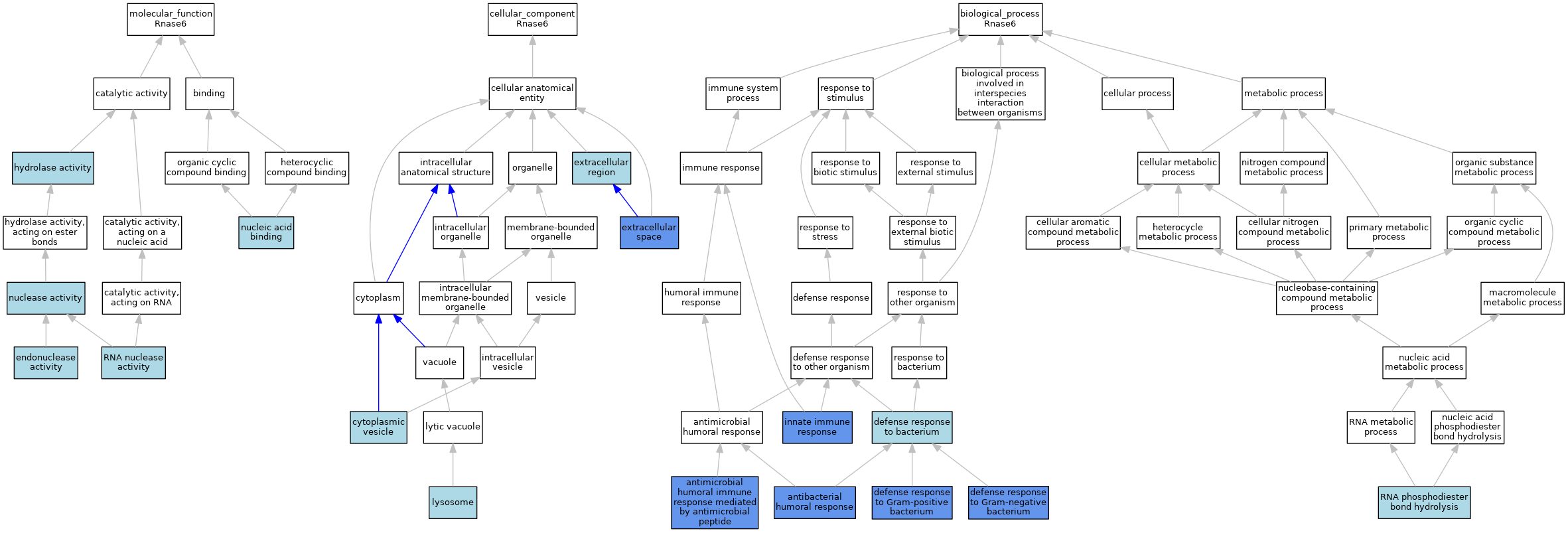
| Molecular Function | GO:0004519 | endonuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004518 | nuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004540 | RNA nuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004540 | RNA nuclease activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:241470 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IEA | J:60000 | |||||||||
| Biological Process | GO:0019731 | antibacterial humoral response | ISO | J:164563 | |||||||||
| Biological Process | GO:0019731 | antibacterial humoral response | IDA | J:241470 | |||||||||
| Biological Process | GO:0019731 | antibacterial humoral response | IBA | J:265628 | |||||||||
| Biological Process | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | IDA | J:241470 | |||||||||
| Biological Process | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | ISO | J:164563 | |||||||||
| Biological Process | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | IBA | J:265628 | |||||||||
| Biological Process | GO:0042742 | defense response to bacterium | IEA | J:60000 | |||||||||
| Biological Process | GO:0050829 | defense response to Gram-negative bacterium | IDA | J:241470 | |||||||||
| Biological Process | GO:0050829 | defense response to Gram-negative bacterium | ISO | J:164563 | |||||||||
| Biological Process | GO:0050829 | defense response to Gram-negative bacterium | IBA | J:265628 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | ISO | J:164563 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | IDA | J:241470 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | IBA | J:265628 | |||||||||
| Biological Process | GO:0045087 | innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IDA | J:241470 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0090501 | RNA phosphodiester bond hydrolysis | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||