|
Symbol Name ID |
Atg16l1
autophagy related 16 like 1 MGI:1924290 |
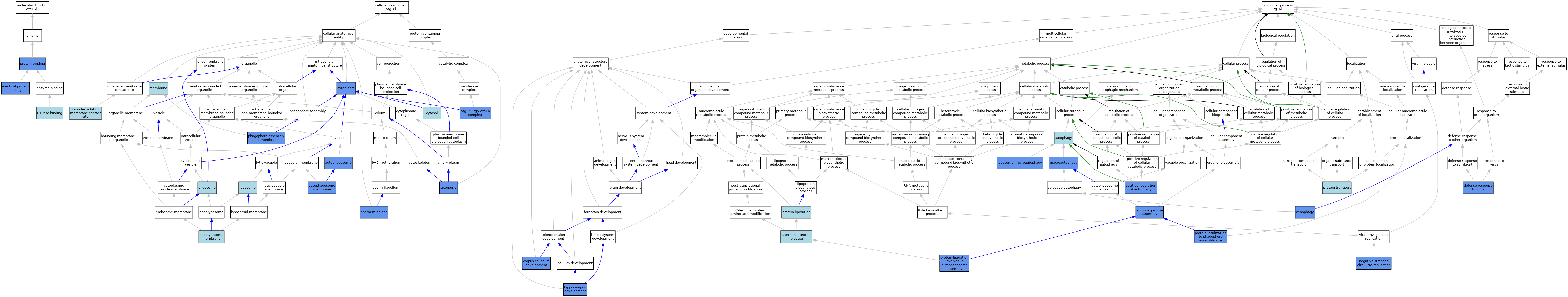
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0051020 | GTPase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:83346 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:319693 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:190086 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245272 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245290 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:203423 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200458 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:215480 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:215480 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:83346 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:83346 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:83346 | |||||||||
| Cellular Component | GO:0034274 | Atg12-Atg5-Atg16 complex | IDA | J:83346 | |||||||||
| Cellular Component | GO:0034274 | Atg12-Atg5-Atg16 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005776 | autophagosome | IDA | J:83346 | |||||||||
| Cellular Component | GO:0000421 | autophagosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000421 | autophagosome membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000421 | autophagosome membrane | IDA | J:167845 | |||||||||
| Cellular Component | GO:0005930 | axoneme | IDA | J:203423 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:319693 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5682677 | |||||||||
| Cellular Component | GO:0036020 | endolysosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0034045 | phagophore assembly site membrane | IDA | J:83346 | |||||||||
| Cellular Component | GO:0097225 | sperm midpiece | IDA | J:319693 | |||||||||
| Cellular Component | GO:0120095 | vacuole-isolation membrane contact site | ISO | J:164563 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IMP | J:70539 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | TAS | J:83346 | |||||||||
| Biological Process | GO:0006914 | autophagy | IEA | J:60000 | |||||||||
| Biological Process | GO:0006501 | C-terminal protein lipidation | ISO | J:164563 | |||||||||
| Biological Process | GO:0022038 | corpus callosum development | IMP | J:249035 | |||||||||
| Biological Process | GO:0051607 | defense response to virus | IMP | J:308892 | |||||||||
| Biological Process | GO:0021766 | hippocampus development | IMP | J:249035 | |||||||||
| Biological Process | GO:0016237 | lysosomal microautophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0016237 | lysosomal microautophagy | IMP | J:308892 | |||||||||
| Biological Process | GO:0016236 | macroautophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0016236 | macroautophagy | IMP | J:70539 | |||||||||
| Biological Process | GO:0039689 | negative stranded viral RNA replication | IBA | J:265628 | |||||||||
| Biological Process | GO:0039689 | negative stranded viral RNA replication | IMP | J:206800 | |||||||||
| Biological Process | GO:0010508 | positive regulation of autophagy | IDA | J:268019 | |||||||||
| Biological Process | GO:0006497 | protein lipidation | ISO | J:164563 | |||||||||
| Biological Process | GO:0061739 | protein lipidation involved in autophagosome assembly | IDA | J:215480 | |||||||||
| Biological Process | GO:0034497 | protein localization to phagophore assembly site | IPI | J:215480 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0098792 | xenophagy | IPI | J:215480 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||