|
Symbol Name ID |
Jade2
jade family PHD finger 2 MGI:1924151 |
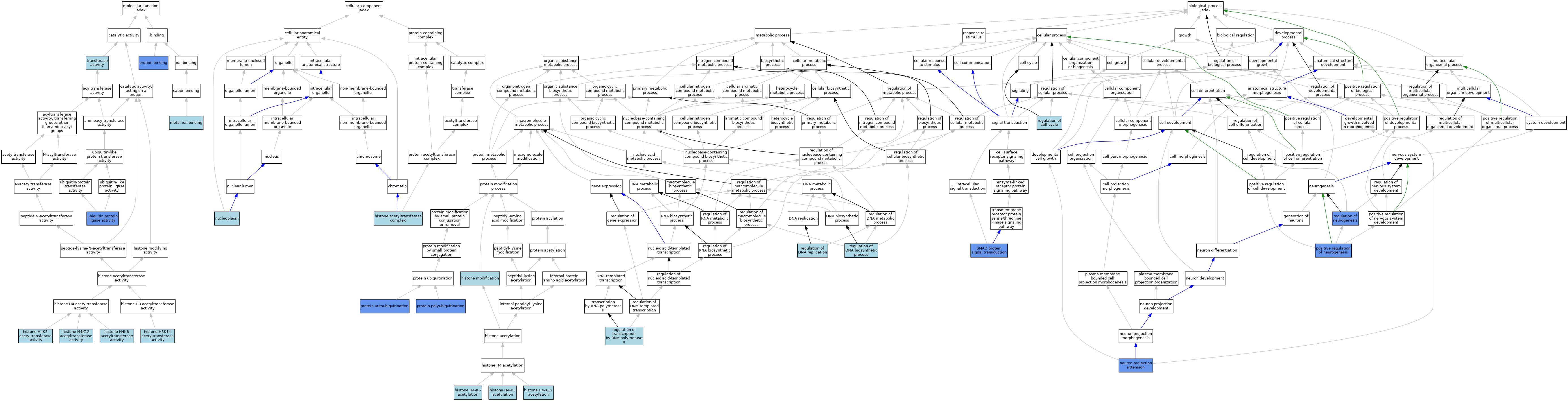
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0036408 | histone H3K14 acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043997 | histone H4K12 acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043995 | histone H4K5 acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043996 | histone H4K8 acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:215624 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0061630 | ubiquitin protein ligase activity | IDA | J:215624 | |||||||||
| Cellular Component | GO:0000123 | histone acetyltransferase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000123 | histone acetyltransferase complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Biological Process | GO:0043983 | histone H4-K12 acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043981 | histone H4-K5 acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043982 | histone H4-K8 acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0016570 | histone modification | ISO | J:164563 | |||||||||
| Biological Process | GO:1990138 | neuron projection extension | IGI | J:215624 | |||||||||
| Biological Process | GO:0050769 | positive regulation of neurogenesis | IMP | J:215624 | |||||||||
| Biological Process | GO:0050769 | positive regulation of neurogenesis | IDA | J:215624 | |||||||||
| Biological Process | GO:0051865 | protein autoubiquitination | IDA | J:215624 | |||||||||
| Biological Process | GO:0000209 | protein polyubiquitination | IMP | J:215624 | |||||||||
| Biological Process | GO:0000209 | protein polyubiquitination | IDA | J:215624 | |||||||||
| Biological Process | GO:0051726 | regulation of cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:2000278 | regulation of DNA biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006275 | regulation of DNA replication | ISO | J:164563 | |||||||||
| Biological Process | GO:0050767 | regulation of neurogenesis | IGI | J:215624 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0060395 | SMAD protein signal transduction | IDA | J:195995 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||