|
Symbol Name ID |
Ush1c
USH1 protein network component harmonin MGI:1919338 |
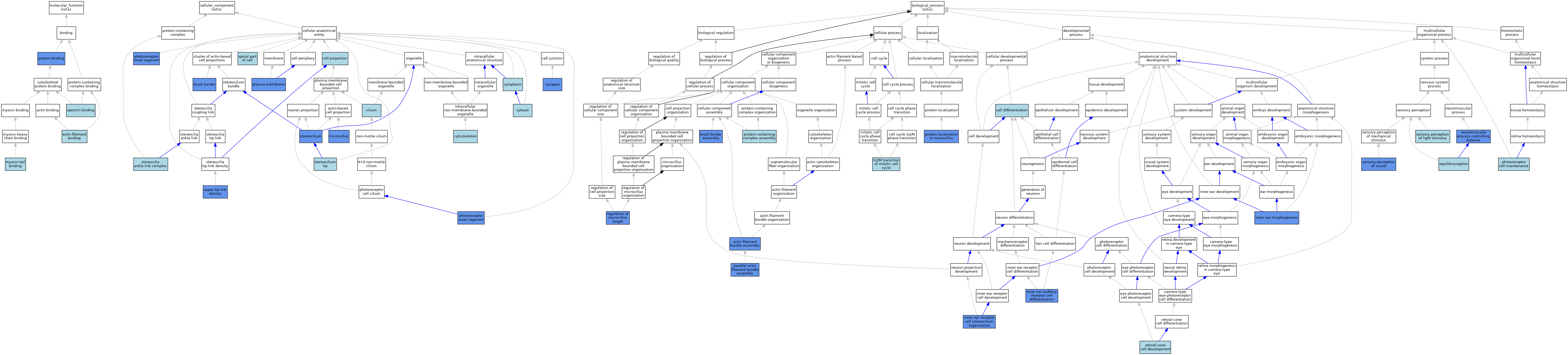
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0051015 | actin filament binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0032029 | myosin tail binding | ISO | J:199579 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:106804 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:99757 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:186070 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:95929 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:80836 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:95929 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:95929 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:82099 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:105750 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:80836 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:95929 | |||||||||
| Molecular Function | GO:0030507 | spectrin binding | ISO | J:199579 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005903 | brush border | IDA | J:212327 | |||||||||
| Cellular Component | GO:0005903 | brush border | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005903 | brush border | IDA | J:219622 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005929 | cilium | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9659536 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9663360 | |||||||||
| Cellular Component | GO:0005902 | microvillus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005902 | microvillus | IDA | J:212327 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | IBA | J:265628 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | IDA | J:86248 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | IDA | J:132040 | |||||||||
| Cellular Component | GO:0001750 | photoreceptor outer segment | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001750 | photoreceptor outer segment | IDA | J:86248 | |||||||||
| Cellular Component | GO:0001750 | photoreceptor outer segment | IDA | J:132040 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:82099 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:82099 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:82099 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:82099 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:82099 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:82099 | |||||||||
| Cellular Component | GO:0002142 | stereocilia ankle link complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | ISO | J:155856 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | EXP | J:230827 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | IDA | J:80836 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | IDA | J:80836 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | IDA | J:80836 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | IDA | J:157099 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | IDA | J:80836 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | IDA | J:80836 | |||||||||
| Cellular Component | GO:0032420 | stereocilium | IDA | J:80836 | |||||||||
| Cellular Component | GO:0032426 | stereocilium tip | IBA | J:265628 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:86248 | |||||||||
| Cellular Component | GO:1990435 | upper tip-link density | IDA | J:283627 | |||||||||
| Biological Process | GO:0051017 | actin filament bundle assembly | IDA | J:80836 | |||||||||
| Biological Process | GO:1904970 | brush border assembly | IMP | J:212327 | |||||||||
| Biological Process | GO:1904970 | brush border assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0050957 | equilibrioception | ISO | J:164563 | |||||||||
| Biological Process | GO:0000086 | G2/M transition of mitotic cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0042491 | inner ear auditory receptor cell differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0042491 | inner ear auditory receptor cell differentiation | IMP | J:85400 | |||||||||
| Biological Process | GO:0042491 | inner ear auditory receptor cell differentiation | IMP | J:85400 | |||||||||
| Biological Process | GO:0042472 | inner ear morphogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0042472 | inner ear morphogenesis | IMP | J:85400 | |||||||||
| Biological Process | GO:0042472 | inner ear morphogenesis | IMP | J:85400 | |||||||||
| Biological Process | GO:0060122 | inner ear receptor cell stereocilium organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0060122 | inner ear receptor cell stereocilium organization | IMP | J:135991 | |||||||||
| Biological Process | GO:0050885 | neuromuscular process controlling balance | IMP | J:85400 | |||||||||
| Biological Process | GO:0050885 | neuromuscular process controlling balance | IMP | J:85400 | |||||||||
| Biological Process | GO:0030046 | parallel actin filament bundle assembly | IDA | J:80836 | |||||||||
| Biological Process | GO:0045494 | photoreceptor cell maintenance | ISO | J:164563 | |||||||||
| Biological Process | GO:1904106 | protein localization to microvillus | ISO | J:164563 | |||||||||
| Biological Process | GO:1904106 | protein localization to microvillus | IMP | J:212327 | |||||||||
| Biological Process | GO:0065003 | protein-containing complex assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0032532 | regulation of microvillus length | IMP | J:212327 | |||||||||
| Biological Process | GO:0046549 | retinal cone cell development | IBA | J:265628 | |||||||||
| Biological Process | GO:0050953 | sensory perception of light stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0007605 | sensory perception of sound | ISO | J:164563 | |||||||||
| Biological Process | GO:0007605 | sensory perception of sound | IBA | J:265628 | |||||||||
| Biological Process | GO:0007605 | sensory perception of sound | IMP | J:85400 | |||||||||
| Biological Process | GO:0007605 | sensory perception of sound | IMP | J:85400 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||