|
Symbol Name ID |
Esco2
establishment of sister chromatid cohesion N-acetyltransferase 2 MGI:1919238 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
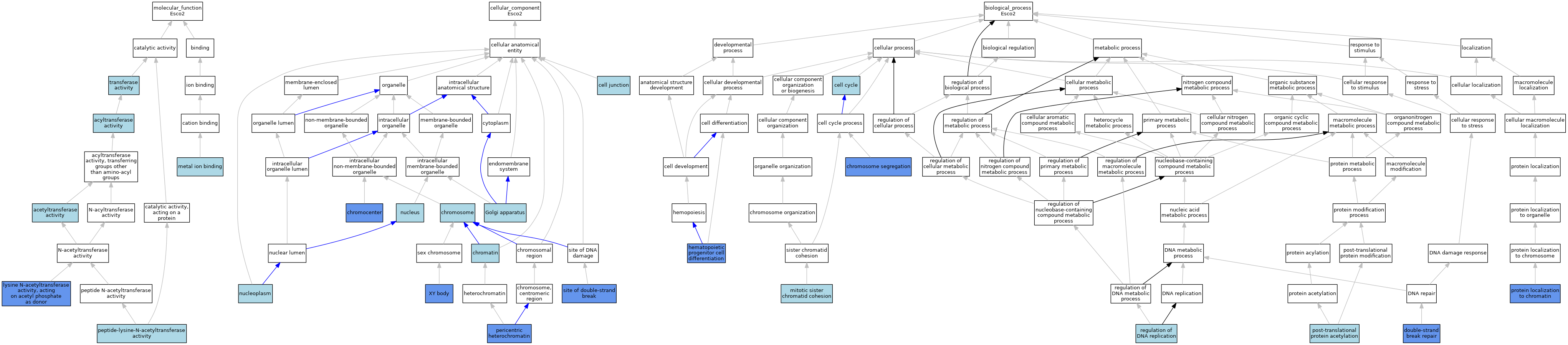
| Molecular Function | GO:0016407 | acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016746 | acyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor | IMP | J:180371 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0061733 | peptide-lysine-N-acetyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030054 | cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000785 | chromatin | IBA | J:265628 | |||||||||
| Cellular Component | GO:0010369 | chromocenter | IDA | J:180371 | |||||||||
| Cellular Component | GO:0005694 | chromosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005721 | pericentric heterochromatin | IDA | J:180371 | |||||||||
| Cellular Component | GO:0035861 | site of double-strand break | IDA | J:197587 | |||||||||
| Cellular Component | GO:0035861 | site of double-strand break | IDA | J:192409 | |||||||||
| Cellular Component | GO:0001741 | XY body | IDA | J:192409 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0007059 | chromosome segregation | IMP | J:180371 | |||||||||
| Biological Process | GO:0006302 | double-strand break repair | IMP | J:197587 | |||||||||
| Biological Process | GO:0002244 | hematopoietic progenitor cell differentiation | IGI | J:202604 | |||||||||
| Biological Process | GO:0007064 | mitotic sister chromatid cohesion | IBA | J:265628 | |||||||||
| Biological Process | GO:0034421 | post-translational protein acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0071168 | protein localization to chromatin | IMP | J:180371 | |||||||||
| Biological Process | GO:0006275 | regulation of DNA replication | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||