|
Symbol Name ID |
Ptk7
PTK7 protein tyrosine kinase 7 MGI:1918711 |
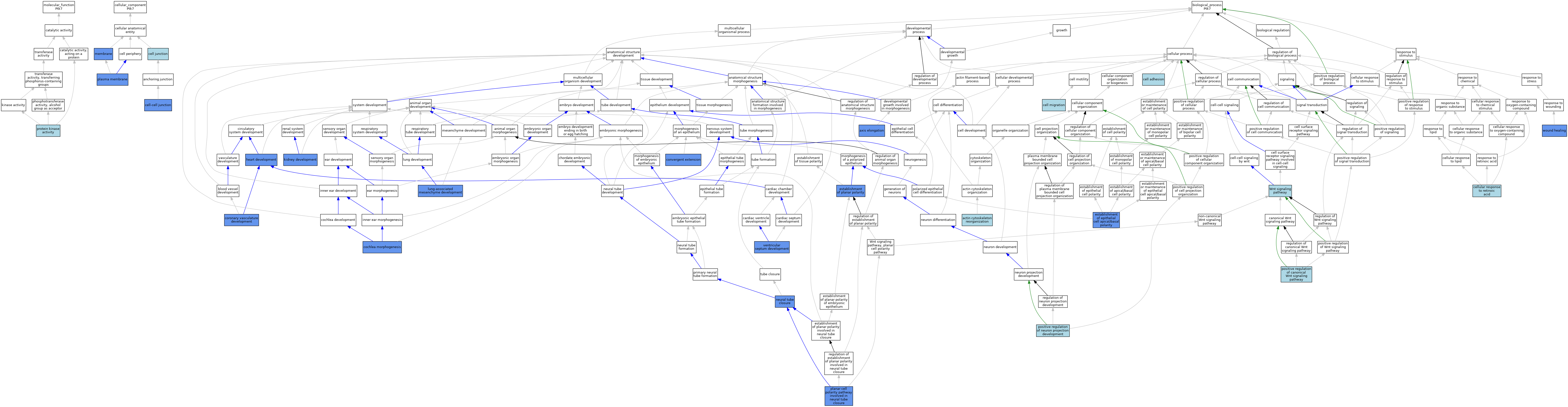
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004672 | protein kinase activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | IDA | J:158680 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:163834 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:158680 | |||||||||
| Biological Process | GO:0031532 | actin cytoskeleton reorganization | ISO | J:164563 | |||||||||
| Biological Process | GO:0003401 | axis elongation | IMP | J:163834 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IEA | J:60000 | |||||||||
| Biological Process | GO:0016477 | cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0071300 | cellular response to retinoic acid | ISO | J:164563 | |||||||||
| Biological Process | GO:0090103 | cochlea morphogenesis | IMP | J:163834 | |||||||||
| Biological Process | GO:0060026 | convergent extension | IMP | J:163834 | |||||||||
| Biological Process | GO:0060976 | coronary vasculature development | IMP | J:222159 | |||||||||
| Biological Process | GO:0045198 | establishment of epithelial cell apical/basal polarity | IMP | J:91298 | |||||||||
| Biological Process | GO:0001736 | establishment of planar polarity | IMP | J:163834 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:222159 | |||||||||
| Biological Process | GO:0001822 | kidney development | IMP | J:231949 | |||||||||
| Biological Process | GO:0060484 | lung-associated mesenchyme development | IMP | J:163834 | |||||||||
| Biological Process | GO:0001843 | neural tube closure | IGI | J:91298 | |||||||||
| Biological Process | GO:0001843 | neural tube closure | IMP | J:163834 | |||||||||
| Biological Process | GO:0001843 | neural tube closure | IMP | J:91298 | |||||||||
| Biological Process | GO:0090179 | planar cell polarity pathway involved in neural tube closure | IGI | J:163834 | |||||||||
| Biological Process | GO:0090179 | planar cell polarity pathway involved in neural tube closure | IGI | J:163834 | |||||||||
| Biological Process | GO:0090263 | positive regulation of canonical Wnt signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0010976 | positive regulation of neuron projection development | ISO | J:164563 | |||||||||
| Biological Process | GO:0003281 | ventricular septum development | IMP | J:222159 | |||||||||
| Biological Process | GO:0016055 | Wnt signaling pathway | IEA | J:60000 | |||||||||
| Biological Process | GO:0042060 | wound healing | IGI | J:162624 | |||||||||
| Biological Process | GO:0042060 | wound healing | IGI | J:162624 | |||||||||
| Biological Process | GO:0042060 | wound healing | IGI | J:162624 | |||||||||
| Biological Process | GO:0042060 | wound healing | IGI | J:162624 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||