|
Symbol Name ID |
Dlg5
discs large MAGUK scaffold protein 5 MGI:1918478 |
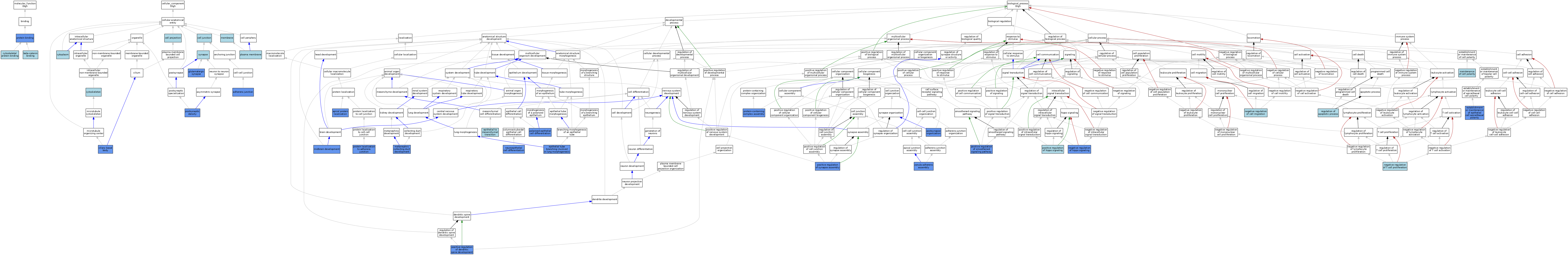
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008013 | beta-catenin binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0008092 | cytoskeletal protein binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:218232 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:238700 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:216499 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:124940 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | IDA | J:124940 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | IDA | J:94763 | |||||||||
| Cellular Component | GO:0030054 | cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0036064 | ciliary basal body | IDA | J:218232 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:216499 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:216499 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IEP | J:216499 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:216499 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:216499 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IMP | J:216499 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0014069 | postsynaptic density | IDA | J:216499 | |||||||||
| Cellular Component | GO:0014069 | postsynaptic density | IEP | J:216499 | |||||||||
| Cellular Component | GO:0014069 | postsynaptic density | IDA | J:216499 | |||||||||
| Cellular Component | GO:0045202 | synapse | IEA | J:60000 | |||||||||
| Biological Process | GO:0045176 | apical protein localization | IMP | J:196288 | |||||||||
| Biological Process | GO:0001837 | epithelial to mesenchymal transition | ISO | J:164563 | |||||||||
| Biological Process | GO:0060441 | epithelial tube branching involved in lung morphogenesis | IMP | J:196288 | |||||||||
| Biological Process | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | IMP | J:196288 | |||||||||
| Biological Process | GO:0030011 | maintenance of cell polarity | ISO | J:164563 | |||||||||
| Biological Process | GO:0072205 | metanephric collecting duct development | IMP | J:124940 | |||||||||
| Biological Process | GO:0030901 | midbrain development | IMP | J:124940 | |||||||||
| Biological Process | GO:0030336 | negative regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0035331 | negative regulation of hippo signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0035331 | negative regulation of hippo signaling | IMP | J:238700 | |||||||||
| Biological Process | GO:0035331 | negative regulation of hippo signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0042130 | negative regulation of T cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0060563 | neuroepithelial cell differentiation | IMP | J:124940 | |||||||||
| Biological Process | GO:0030859 | polarized epithelial cell differentiation | IMP | J:124940 | |||||||||
| Biological Process | GO:0030859 | polarized epithelial cell differentiation | IMP | J:124940 | |||||||||
| Biological Process | GO:0060999 | positive regulation of dendritic spine development | IMP | J:216499 | |||||||||
| Biological Process | GO:0035332 | positive regulation of hippo signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0045880 | positive regulation of smoothened signaling pathway | IMP | J:218232 | |||||||||
| Biological Process | GO:0051965 | positive regulation of synapse assembly | IMP | J:216499 | |||||||||
| Biological Process | GO:0099173 | postsynapse organization | IDA | J:216499 | |||||||||
| Biological Process | GO:0099173 | postsynapse organization | IMP | J:216499 | |||||||||
| Biological Process | GO:0099173 | postsynapse organization | IDA | J:216499 | |||||||||
| Biological Process | GO:0071896 | protein localization to adherens junction | IMP | J:124940 | |||||||||
| Biological Process | GO:0071896 | protein localization to adherens junction | IMP | J:124940 | |||||||||
| Biological Process | GO:0065003 | protein-containing complex assembly | IMP | J:124940 | |||||||||
| Biological Process | GO:0065003 | protein-containing complex assembly | IMP | J:124940 | |||||||||
| Biological Process | GO:0042981 | regulation of apoptotic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0045186 | zonula adherens assembly | IMP | J:124940 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||