|
Symbol Name ID |
Gtpbp4
GTP binding protein 4 MGI:1916487 |
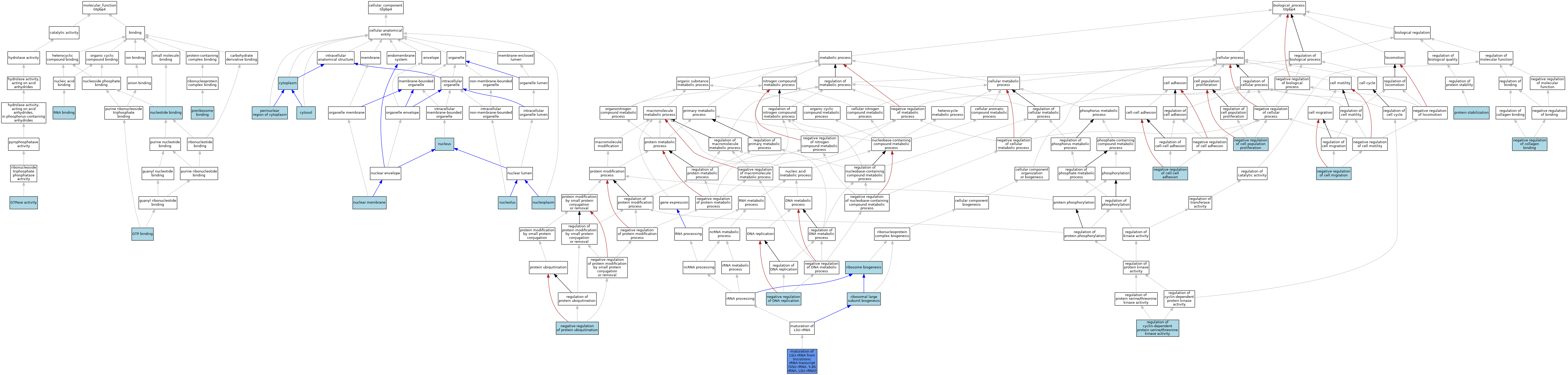
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005525 | GTP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:1990275 | preribosome binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Biological Process | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | IMP | J:218306 | |||||||||
| Biological Process | GO:0030336 | negative regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0022408 | negative regulation of cell-cell adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0033342 | negative regulation of collagen binding | ISO | J:164563 | |||||||||
| Biological Process | GO:0008156 | negative regulation of DNA replication | ISO | J:164563 | |||||||||
| Biological Process | GO:0031397 | negative regulation of protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0042273 | ribosomal large subunit biogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0042273 | ribosomal large subunit biogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0042254 | ribosome biogenesis | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||