|
Symbol Name ID |
Kmt5a
lysine methyltransferase 5A MGI:1915206 |
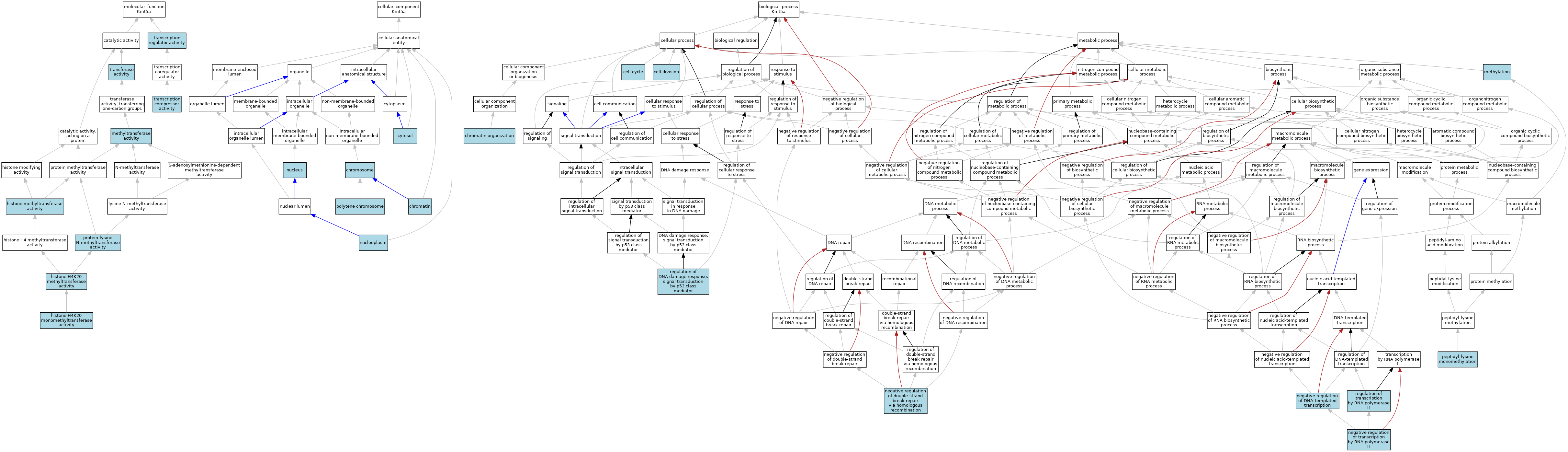
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042799 | histone H4K20 methyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042799 | histone H4K20 methyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0140944 | histone H4K20 monomethyltransferase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0042054 | histone methyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008168 | methyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016279 | protein-lysine N-methyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0140110 | transcription regulator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005700 | polytene chromosome | IBA | J:265628 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0051301 | cell division | IEA | J:60000 | |||||||||
| Biological Process | GO:0006325 | chromatin organization | IEA | J:60000 | |||||||||
| Biological Process | GO:0032259 | methylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0018026 | peptidyl-lysine monomethylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | ISO | J:164563 | |||||||||
| Biological Process | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | IBA | J:265628 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||