|
Symbol Name ID |
Dcxr
dicarbonyl L-xylulose reductase MGI:1915130 |
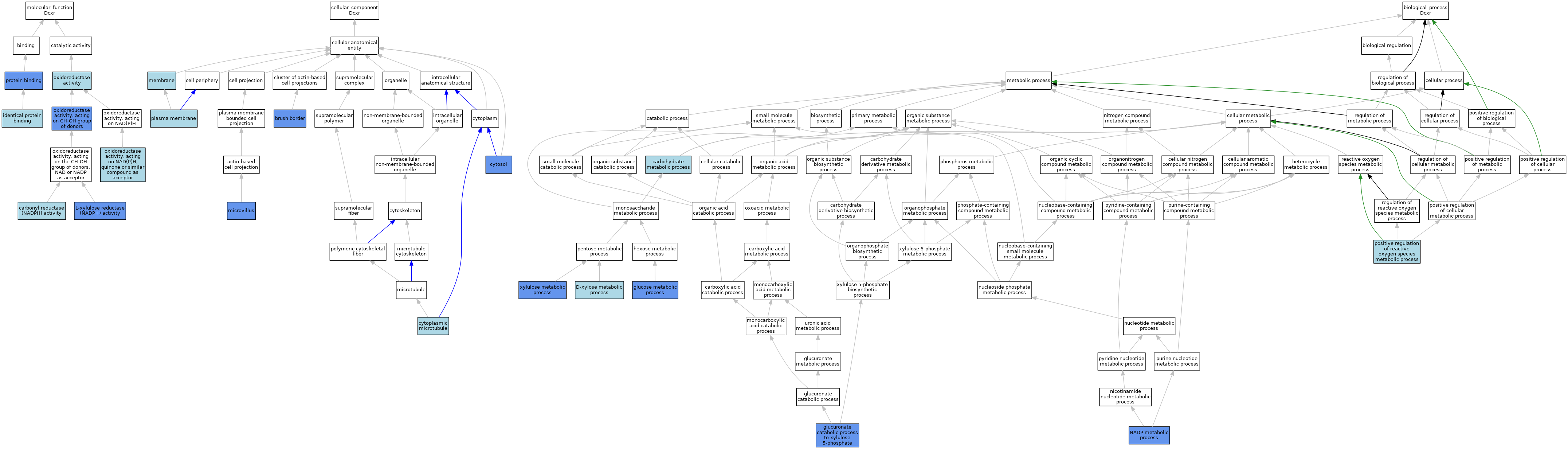
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004090 | carbonyl reductase (NADPH) activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0050038 | L-xylulose reductase (NADP+) activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0050038 | L-xylulose reductase (NADP+) activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0050038 | L-xylulose reductase (NADP+) activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0050038 | L-xylulose reductase (NADP+) activity | IDA | J:76552 | |||||||||
| Molecular Function | GO:0050038 | L-xylulose reductase (NADP+) activity | ISO | J:323238 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | IDA | J:76552 | |||||||||
| Molecular Function | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:76552 | |||||||||
| Cellular Component | GO:0005903 | brush border | IDA | J:76552 | |||||||||
| Cellular Component | GO:0005881 | cytoplasmic microtubule | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:323240 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005902 | microvillus | IDA | J:76552 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0005975 | carbohydrate metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0042732 | D-xylose metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006006 | glucose metabolic process | IDA | J:76552 | |||||||||
| Biological Process | GO:0006006 | glucose metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006006 | glucose metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006006 | glucose metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0019640 | glucuronate catabolic process to xylulose 5-phosphate | IDA | J:76552 | |||||||||
| Biological Process | GO:0019640 | glucuronate catabolic process to xylulose 5-phosphate | ISO | J:323238 | |||||||||
| Biological Process | GO:0006739 | NADP metabolic process | IDA | J:76552 | |||||||||
| Biological Process | GO:2000379 | positive regulation of reactive oxygen species metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0005997 | xylulose metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0005997 | xylulose metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0005997 | xylulose metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0005997 | xylulose metabolic process | IDA | J:76552 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||