|
Symbol Name ID |
Wdr48
WD repeat domain 48 MGI:1914811 |
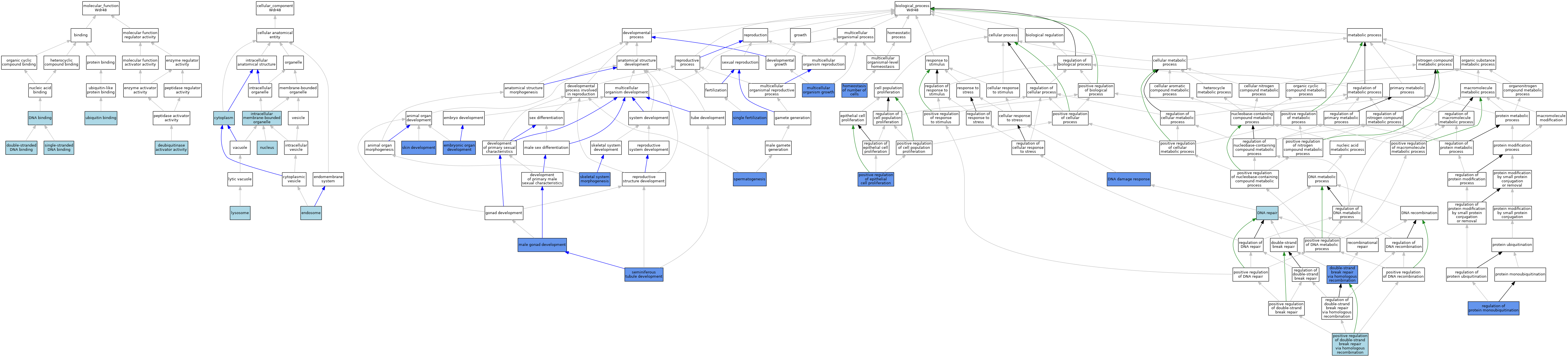
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0035800 | deubiquitinase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003690 | double-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003697 | single-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043130 | ubiquitin binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IMP | J:206106 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:60000 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | IBA | J:265628 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | IMP | J:206106 | |||||||||
| Biological Process | GO:0048568 | embryonic organ development | IMP | J:206106 | |||||||||
| Biological Process | GO:0048872 | homeostasis of number of cells | IMP | J:206106 | |||||||||
| Biological Process | GO:0008584 | male gonad development | IMP | J:206106 | |||||||||
| Biological Process | GO:0035264 | multicellular organism growth | IMP | J:206106 | |||||||||
| Biological Process | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0050679 | positive regulation of epithelial cell proliferation | IMP | J:206106 | |||||||||
| Biological Process | GO:1902525 | regulation of protein monoubiquitination | IMP | J:206106 | |||||||||
| Biological Process | GO:0072520 | seminiferous tubule development | IMP | J:206106 | |||||||||
| Biological Process | GO:0007338 | single fertilization | IMP | J:206106 | |||||||||
| Biological Process | GO:0048705 | skeletal system morphogenesis | IMP | J:206106 | |||||||||
| Biological Process | GO:0043588 | skin development | IMP | J:206106 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:206106 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||