|
Symbol Name ID |
Rnf186
ring finger protein 186 MGI:1914075 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
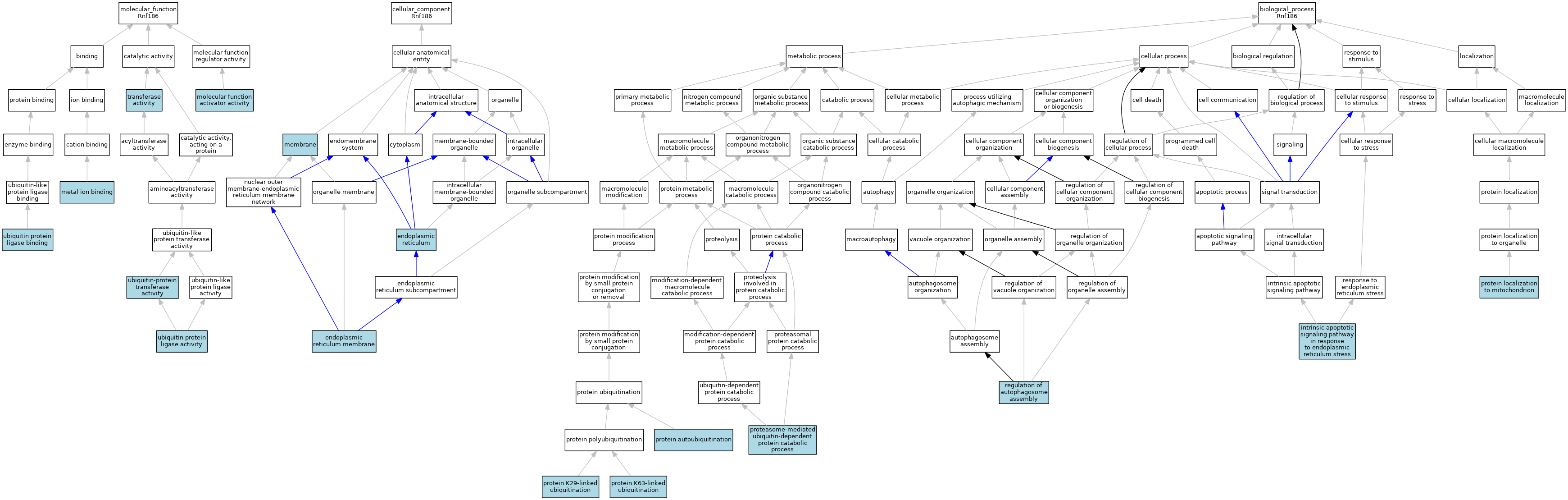
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0140677 | molecular function activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0061630 | ubiquitin protein ligase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0061630 | ubiquitin protein ligase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004842 | ubiquitin-protein transferase activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | ISO | J:164563 | |||||||||
| Biological Process | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | IBA | J:265628 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0051865 | protein autoubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0051865 | protein autoubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0035519 | protein K29-linked ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0035519 | protein K29-linked ubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0070534 | protein K63-linked ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0070534 | protein K63-linked ubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0070585 | protein localization to mitochondrion | ISO | J:164563 | |||||||||
| Biological Process | GO:0070585 | protein localization to mitochondrion | IBA | J:265628 | |||||||||
| Biological Process | GO:2000785 | regulation of autophagosome assembly | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||