|
Symbol Name ID |
Foxo4
forkhead box O4 MGI:1891915 |
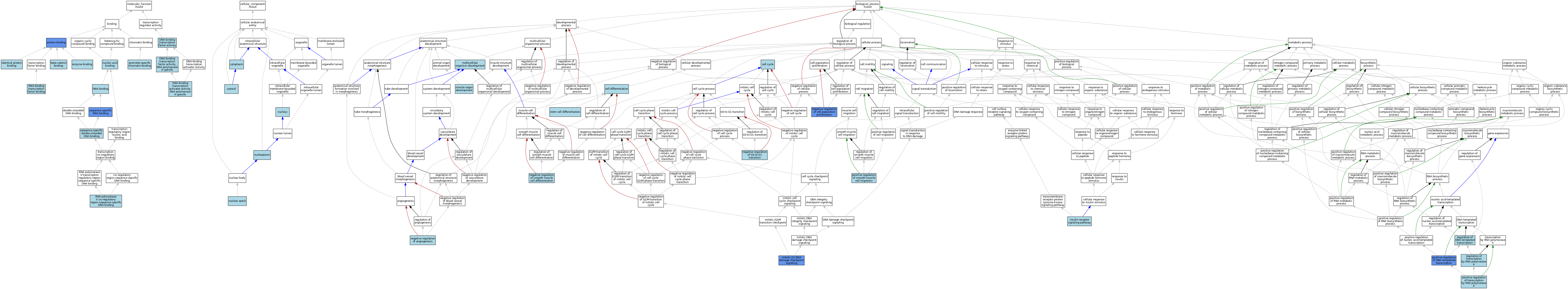
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008013 | beta-catenin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0140297 | DNA-binding transcription factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990841 | promoter-specific chromatin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200381 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043565 | sequence-specific DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043565 | sequence-specific DNA binding | IDA | J:69574 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9617657 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9617711 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9617755 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-NUL-9617710 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-NUL-9617742 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-NUL-9617757 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-NUL-9617903 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0008286 | insulin receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | IDA | J:78043 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0007517 | muscle organ development | IEA | J:60000 | |||||||||
| Biological Process | GO:0016525 | negative regulation of angiogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | IDA | J:78043 | |||||||||
| Biological Process | GO:0070317 | negative regulation of G0 to G1 transition | ISO | J:164563 | |||||||||
| Biological Process | GO:0051151 | negative regulation of smooth muscle cell differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IDA | J:88189 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IDA | J:78043 | |||||||||
| Biological Process | GO:0014911 | positive regulation of smooth muscle cell migration | ISO | J:155856 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0048863 | stem cell differentiation | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||