|
Symbol Name ID |
Fzd2
frizzled class receptor 2 MGI:1888513 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
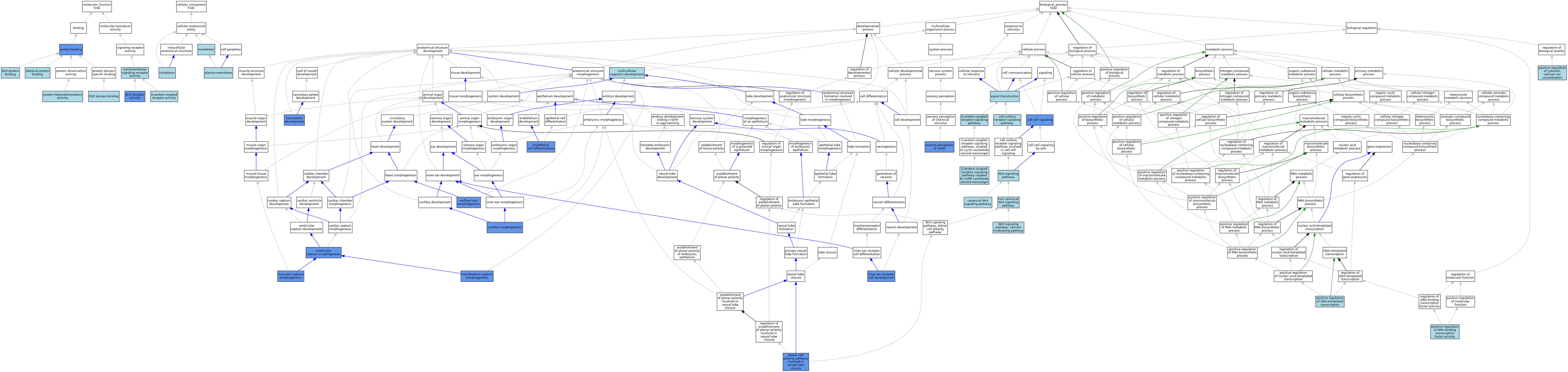
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0030165 | PDZ domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:323230 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:89972 | |||||||||
| Molecular Function | GO:0046982 | protein heterodimerization activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004888 | transmembrane signaling receptor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0042813 | Wnt receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042813 | Wnt receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042813 | Wnt receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0042813 | Wnt receptor activity | IDA | J:161589 | |||||||||
| Molecular Function | GO:0017147 | Wnt-protein binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0060070 | canonical Wnt signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0060070 | canonical Wnt signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0060070 | canonical Wnt signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007166 | cell surface receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0007267 | cell-cell signaling | IDA | J:98305 | |||||||||
| Biological Process | GO:0007267 | cell-cell signaling | IDA | J:98305 | |||||||||
| Biological Process | GO:0090103 | cochlea morphogenesis | IGI | J:165556 | |||||||||
| Biological Process | GO:0045446 | endothelial cell differentiation | IDA | J:98305 | |||||||||
| Biological Process | GO:0045446 | endothelial cell differentiation | IGI | J:98305 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:60000 | |||||||||
| Biological Process | GO:0007199 | G protein-coupled receptor signaling pathway coupled to cGMP nucleotide second messenger | ISO | J:155856 | |||||||||
| Biological Process | GO:0060022 | hard palate development | IGI | J:165556 | |||||||||
| Biological Process | GO:0060022 | hard palate development | IMP | J:165556 | |||||||||
| Biological Process | GO:0060119 | inner ear receptor cell development | IGI | J:165556 | |||||||||
| Biological Process | GO:0003149 | membranous septum morphogenesis | IGI | J:165556 | |||||||||
| Biological Process | GO:0003149 | membranous septum morphogenesis | IGI | J:165556 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0003150 | muscular septum morphogenesis | IGI | J:165556 | |||||||||
| Biological Process | GO:0003150 | muscular septum morphogenesis | IGI | J:165556 | |||||||||
| Biological Process | GO:0035567 | non-canonical Wnt signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0003151 | outflow tract morphogenesis | IGI | J:165556 | |||||||||
| Biological Process | GO:0090179 | planar cell polarity pathway involved in neural tube closure | IGI | J:165556 | |||||||||
| Biological Process | GO:0090179 | planar cell polarity pathway involved in neural tube closure | IGI | J:165556 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | ISO | J:155856 | |||||||||
| Biological Process | GO:0051091 | positive regulation of DNA-binding transcription factor activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0007608 | sensory perception of smell | IMP | J:165556 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0060412 | ventricular septum morphogenesis | IGI | J:165556 | |||||||||
| Biological Process | GO:0016055 | Wnt signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||