|
Symbol Name ID |
Clec4n
C-type lectin domain family 4, member n MGI:1861231 |
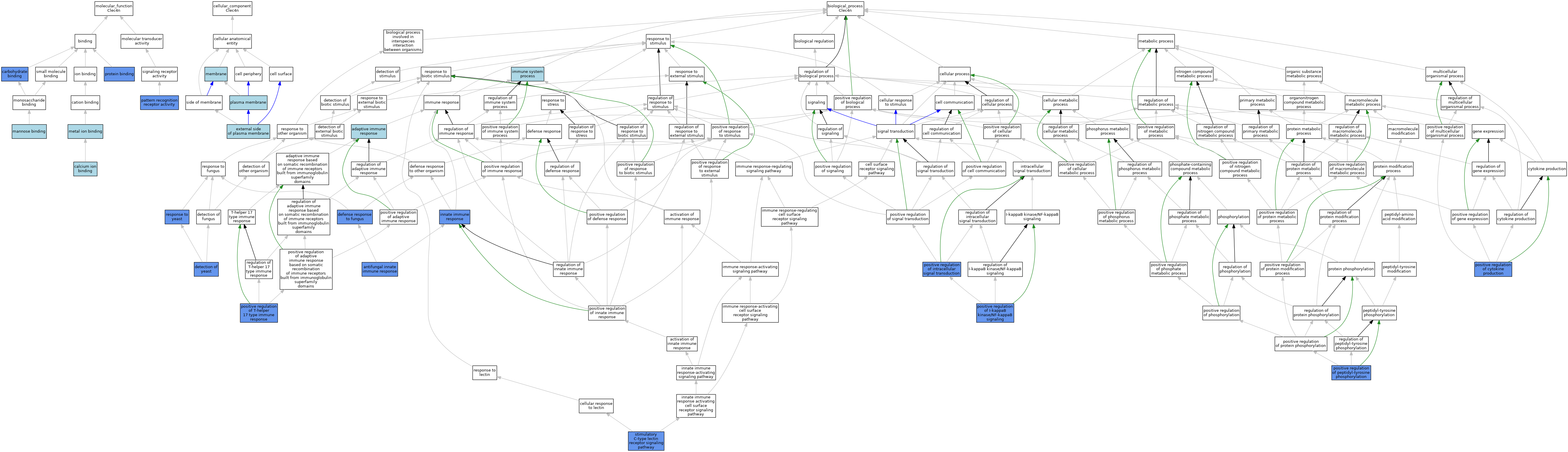
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030246 | carbohydrate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030246 | carbohydrate binding | IDA | J:160680 | |||||||||
| Molecular Function | GO:0030246 | carbohydrate binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005537 | mannose binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0038187 | pattern recognition receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0038187 | pattern recognition receptor activity | IMP | J:178744 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:168200 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-5621348 | |||||||||
| Biological Process | GO:0002250 | adaptive immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0061760 | antifungal innate immune response | IMP | J:291457 | |||||||||
| Biological Process | GO:0061760 | antifungal innate immune response | IMP | J:178744 | |||||||||
| Biological Process | GO:0061760 | antifungal innate immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0050832 | defense response to fungus | IDA | J:160680 | |||||||||
| Biological Process | GO:0001879 | detection of yeast | IMP | J:178744 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IDA | J:168200 | |||||||||
| Biological Process | GO:0001819 | positive regulation of cytokine production | IDA | J:160680 | |||||||||
| Biological Process | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | IDA | J:168200 | |||||||||
| Biological Process | GO:1902533 | positive regulation of intracellular signal transduction | IMP | J:178744 | |||||||||
| Biological Process | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | IMP | J:178744 | |||||||||
| Biological Process | GO:2000318 | positive regulation of T-helper 17 type immune response | IMP | J:291457 | |||||||||
| Biological Process | GO:0001878 | response to yeast | IMP | J:178744 | |||||||||
| Biological Process | GO:0002223 | stimulatory C-type lectin receptor signaling pathway | IMP | J:178744 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||