|
Symbol Name ID |
Mad2l1
MAD2 mitotic arrest deficient-like 1 MGI:1860374 |
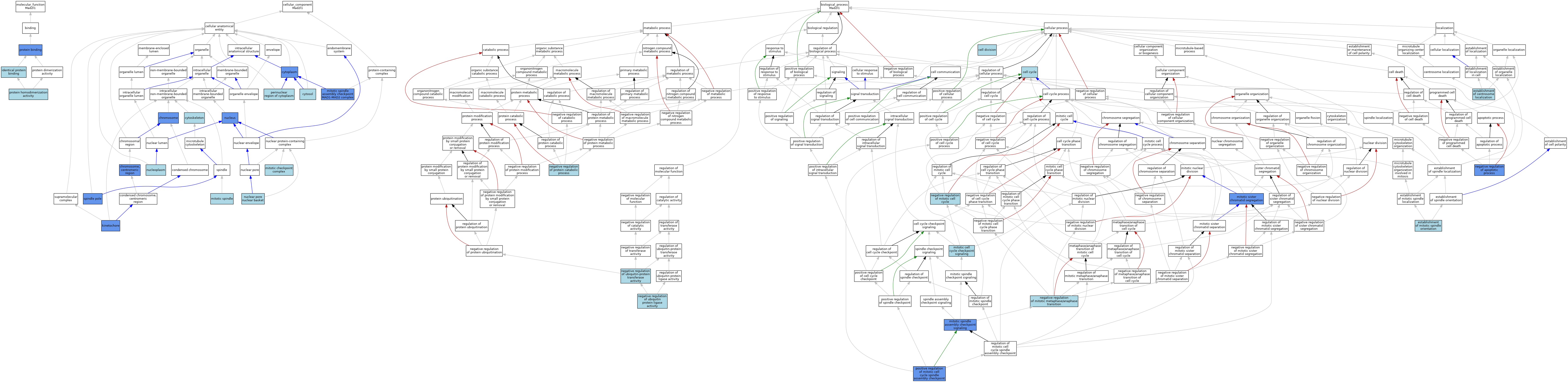
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:305154 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200494 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IDA | J:121535 | |||||||||
| Cellular Component | GO:0000775 | chromosome, centromeric region | IDA | J:80450 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:121560 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | IDA | J:145744 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | IDA | J:148330 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | IDA | J:121560 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | IDA | J:64328 | |||||||||
| Cellular Component | GO:0033597 | mitotic checkpoint complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0072686 | mitotic spindle | ISO | J:164563 | |||||||||
| Cellular Component | GO:1990728 | mitotic spindle assembly checkpoint MAD1-MAD2 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:1990728 | mitotic spindle assembly checkpoint MAD1-MAD2 complex | IPI | J:314961 | |||||||||
| Cellular Component | GO:0044615 | nuclear pore nuclear basket | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:240155 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000922 | spindle pole | IDA | J:121560 | |||||||||
| Cellular Component | GO:0000922 | spindle pole | IDA | J:64328 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0051301 | cell division | IEA | J:60000 | |||||||||
| Biological Process | GO:0051660 | establishment of centrosome localization | ISO | J:164563 | |||||||||
| Biological Process | GO:0000132 | establishment of mitotic spindle orientation | ISO | J:164563 | |||||||||
| Biological Process | GO:0007093 | mitotic cell cycle checkpoint signaling | ISO | J:73065 | |||||||||
| Biological Process | GO:0000070 | mitotic sister chromatid segregation | IMP | J:62829 | |||||||||
| Biological Process | GO:0007094 | mitotic spindle assembly checkpoint signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0007094 | mitotic spindle assembly checkpoint signaling | NAS | J:320291 | |||||||||
| Biological Process | GO:0007094 | mitotic spindle assembly checkpoint signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0007094 | mitotic spindle assembly checkpoint signaling | ISA | J:62829 | |||||||||
| Biological Process | GO:0007094 | mitotic spindle assembly checkpoint signaling | IMP | J:67456 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IMP | J:166080 | |||||||||
| Biological Process | GO:0045930 | negative regulation of mitotic cell cycle | ISO | J:73065 | |||||||||
| Biological Process | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | TAS | J:79594 | |||||||||
| Biological Process | GO:0042177 | negative regulation of protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:1904667 | negative regulation of ubiquitin protein ligase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | ISO | J:164563 | |||||||||
| Biological Process | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | IMP | J:166080 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||