|
Symbol Name ID |
Dll4
delta like canonical Notch ligand 4 MGI:1859388 |
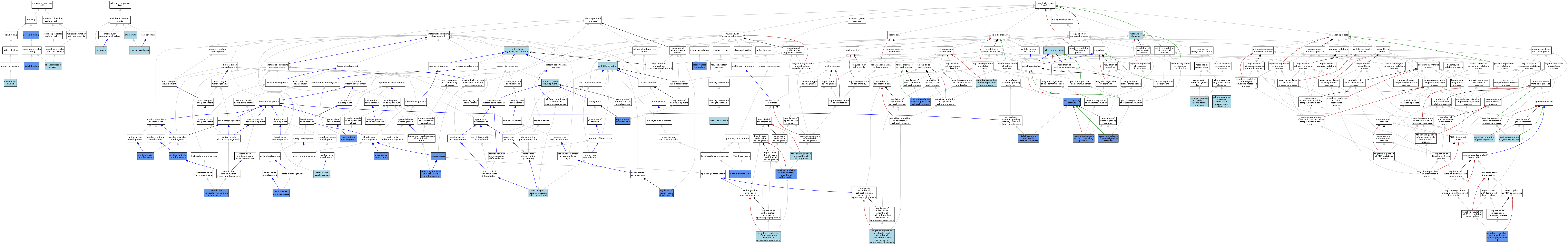
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005112 | Notch binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005112 | Notch binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005112 | Notch binding | IPI | J:167379 | |||||||||
| Molecular Function | GO:0005112 | Notch binding | IDA | J:62507 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:227382 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:101173 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-2053938 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9604278 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISS | J:62507 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IMP | J:93125 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IMP | J:200671 | |||||||||
| Biological Process | GO:0003180 | aortic valve morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0072554 | blood vessel lumenization | IMP | J:93125 | |||||||||
| Biological Process | GO:0001974 | blood vessel remodeling | IMP | J:93125 | |||||||||
| Biological Process | GO:0001569 | branching involved in blood vessel morphogenesis | IMP | J:93157 | |||||||||
| Biological Process | GO:0003209 | cardiac atrium morphogenesis | IMP | J:93157 | |||||||||
| Biological Process | GO:0003208 | cardiac ventricle morphogenesis | IMP | J:93157 | |||||||||
| Biological Process | GO:0007154 | cell communication | IEA | J:72247 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0044344 | cellular response to fibroblast growth factor stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0035912 | dorsal aorta morphogenesis | IMP | J:93157 | |||||||||
| Biological Process | GO:0035912 | dorsal aorta morphogenesis | IMP | J:93125 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:72247 | |||||||||
| Biological Process | GO:0043537 | negative regulation of blood vessel endothelial cell migration | IDA | J:138721 | |||||||||
| Biological Process | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0010596 | negative regulation of endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:0045746 | negative regulation of Notch signaling pathway | IDA | J:224861 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IMP | J:93157 | |||||||||
| Biological Process | GO:0007399 | nervous system development | IEA | J:60000 | |||||||||
| Biological Process | GO:0061314 | Notch signaling involved in heart development | IMP | J:93157 | |||||||||
| Biological Process | GO:0007219 | Notch signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007219 | Notch signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007219 | Notch signaling pathway | IDA | J:212472 | |||||||||
| Biological Process | GO:0003344 | pericardium morphogenesis | IMP | J:93157 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:2000179 | positive regulation of neural precursor cell proliferation | IMP | J:181993 | |||||||||
| Biological Process | GO:0045747 | positive regulation of Notch signaling pathway | IDA | J:224861 | |||||||||
| Biological Process | GO:0061074 | regulation of neural retina development | IMP | J:181993 | |||||||||
| Biological Process | GO:0050767 | regulation of neurogenesis | IMP | J:149456 | |||||||||
| Biological Process | GO:0050896 | response to stimulus | IEA | J:60000 | |||||||||
| Biological Process | GO:0030217 | T cell differentiation | IDA | J:90630 | |||||||||
| Biological Process | GO:0060579 | ventral spinal cord interneuron fate commitment | ISO | J:164563 | |||||||||
| Biological Process | GO:0003222 | ventricular trabecula myocardium morphogenesis | IMP | J:93157 | |||||||||
| Biological Process | GO:0007601 | visual perception | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||