|
Symbol Name ID |
Aldh1a1
aldehyde dehydrogenase family 1, subfamily A1 MGI:1353450 |
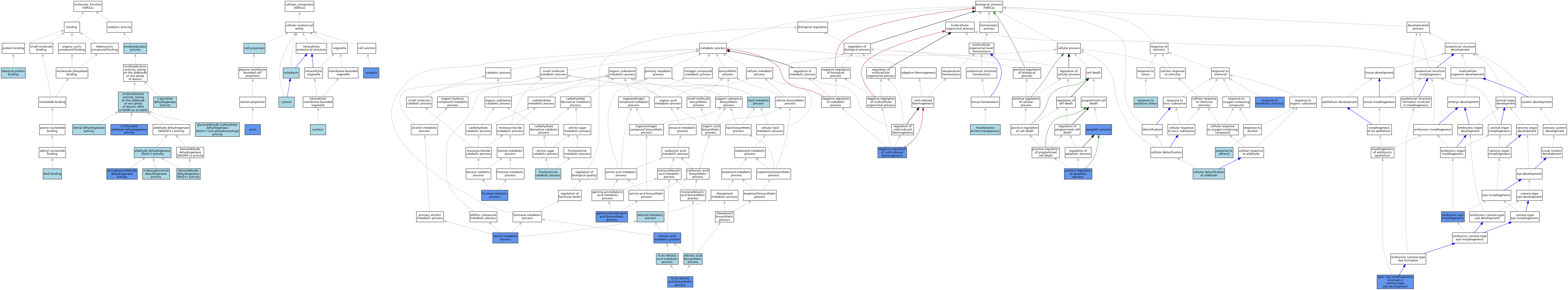
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0033737 | 1-pyrroline dehydrogenase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity | IDA | J:82902 | |||||||||
| Molecular Function | GO:0106373 | 3-deoxyglucosone dehydrogenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004029 | aldehyde dehydrogenase (NAD+) activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004029 | aldehyde dehydrogenase (NAD+) activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004029 | aldehyde dehydrogenase (NAD+) activity | ISO | J:322155 | |||||||||
| Molecular Function | GO:0019145 | aminobutyraldehyde dehydrogenase activity | IMP | J:226538 | |||||||||
| Molecular Function | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0051287 | NAD binding | ISS | J:240299 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | IEA | J:72247 | |||||||||
| Molecular Function | GO:0001758 | retinal dehydrogenase activity | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:226538 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:226538 | |||||||||
| Biological Process | GO:0042904 | 9-cis-retinoic acid biosynthetic process | IDA | J:83534 | |||||||||
| Biological Process | GO:0042905 | 9-cis-retinoic acid metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IGI | J:102847 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IGI | J:108515 | |||||||||
| Biological Process | GO:0110095 | cellular detoxification of aldehyde | ISO | J:164563 | |||||||||
| Biological Process | GO:0110095 | cellular detoxification of aldehyde | ISO | J:155856 | |||||||||
| Biological Process | GO:0048048 | embryonic eye morphogenesis | IGI | J:108515 | |||||||||
| Biological Process | GO:0048048 | embryonic eye morphogenesis | IGI | J:102847 | |||||||||
| Biological Process | GO:0030392 | fructosamine catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006001 | fructose catabolic process | IDA | J:322167 | |||||||||
| Biological Process | GO:0006001 | fructose catabolic process | IDA | J:322154 | |||||||||
| Biological Process | GO:0009449 | gamma-aminobutyric acid biosynthetic process | IMP | J:226538 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0036438 | maintenance of lens transparency | ISO | J:155856 | |||||||||
| Biological Process | GO:0120163 | negative regulation of cold-induced thermogenesis | IMP | J:187531 | |||||||||
| Biological Process | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | IGI | J:108515 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | IGI | J:108515 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | IGI | J:102847 | |||||||||
| Biological Process | GO:0045471 | response to ethanol | ISO | J:155856 | |||||||||
| Biological Process | GO:0006979 | response to oxidative stress | ISO | J:155856 | |||||||||
| Biological Process | GO:0009410 | response to xenobiotic stimulus | IDA | J:145577 | |||||||||
| Biological Process | GO:0002138 | retinoic acid biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0042573 | retinoic acid metabolic process | IGI | J:102847 | |||||||||
| Biological Process | GO:0042573 | retinoic acid metabolic process | IMP | J:83957 | |||||||||
| Biological Process | GO:0042573 | retinoic acid metabolic process | IMP | J:85538 | |||||||||
| Biological Process | GO:0042573 | retinoic acid metabolic process | IGI | J:102847 | |||||||||
| Biological Process | GO:0042573 | retinoic acid metabolic process | IDA | J:54527 | |||||||||
| Biological Process | GO:0042573 | retinoic acid metabolic process | IMP | J:102847 | |||||||||
| Biological Process | GO:0042573 | retinoic acid metabolic process | IGI | J:102847 | |||||||||
| Biological Process | GO:0001523 | retinoid metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0042572 | retinol metabolic process | IMP | J:85538 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||