|
Symbol Name ID |
Pkd2l1
polycystic kidney disease 2-like 1 MGI:1352448 |
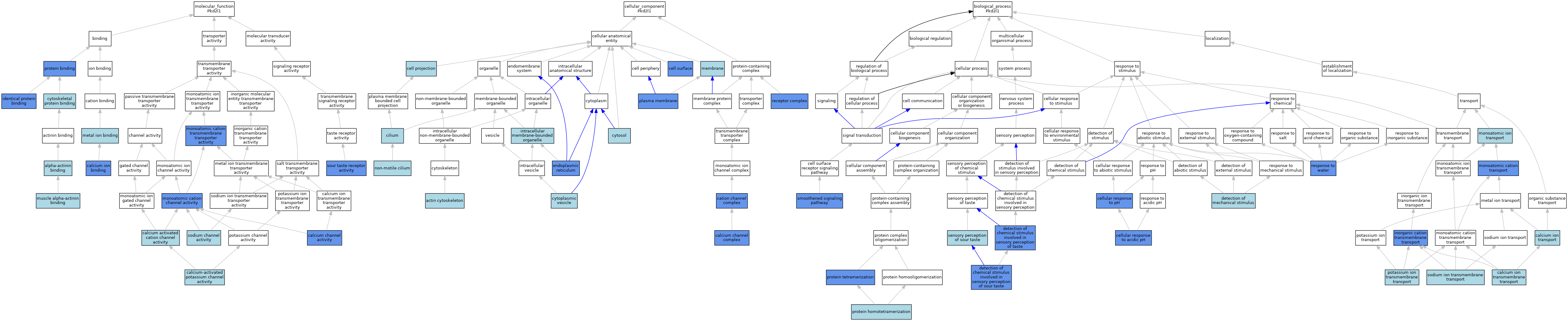
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0051393 | alpha-actinin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005227 | calcium activated cation channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005262 | calcium channel activity | IMP | J:207901 | |||||||||
| Molecular Function | GO:0005262 | calcium channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005262 | calcium channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | IDA | J:182478 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0015269 | calcium-activated potassium channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015269 | calcium-activated potassium channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008092 | cytoskeletal protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:182388 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:182478 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005261 | monoatomic cation channel activity | IDA | J:182388 | |||||||||
| Molecular Function | GO:0005261 | monoatomic cation channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005261 | monoatomic cation channel activity | IDA | J:266474 | |||||||||
| Molecular Function | GO:0005261 | monoatomic cation channel activity | IPI | J:96951 | |||||||||
| Molecular Function | GO:0008324 | monoatomic cation transmembrane transporter activity | IDA | J:182388 | |||||||||
| Molecular Function | GO:0051371 | muscle alpha-actinin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051371 | muscle alpha-actinin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:182388 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:96951 | |||||||||
| Molecular Function | GO:0005272 | sodium channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005272 | sodium channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0033040 | sour taste receptor activity | IDA | J:182388 | |||||||||
| Cellular Component | GO:0015629 | actin cytoskeleton | ISO | J:164563 | |||||||||
| Cellular Component | GO:0034704 | calcium channel complex | IDA | J:283187 | |||||||||
| Cellular Component | GO:0034704 | calcium channel complex | IMP | J:207901 | |||||||||
| Cellular Component | GO:0034704 | calcium channel complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0034703 | cation channel complex | IDA | J:182388 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:182388 | |||||||||
| Cellular Component | GO:0005929 | cilium | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:96951 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0097730 | non-motile cilium | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:266474 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:283187 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IPI | J:96951 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | IDA | J:182388 | |||||||||
| Biological Process | GO:0070588 | calcium ion transmembrane transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0006816 | calcium ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0071468 | cellular response to acidic pH | IDA | J:283187 | |||||||||
| Biological Process | GO:0071468 | cellular response to acidic pH | IDA | J:182388 | |||||||||
| Biological Process | GO:0071467 | cellular response to pH | IDA | J:266474 | |||||||||
| Biological Process | GO:0001581 | detection of chemical stimulus involved in sensory perception of sour taste | IDA | J:182388 | |||||||||
| Biological Process | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste | IMP | J:244208 | |||||||||
| Biological Process | GO:0050982 | detection of mechanical stimulus | IBA | J:265628 | |||||||||
| Biological Process | GO:0098662 | inorganic cation transmembrane transport | IDA | J:266474 | |||||||||
| Biological Process | GO:0098662 | inorganic cation transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006812 | monoatomic cation transport | IDA | J:182388 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0051289 | protein homotetramerization | ISO | J:164563 | |||||||||
| Biological Process | GO:0051262 | protein tetramerization | IDA | J:283187 | |||||||||
| Biological Process | GO:0009415 | response to water | IMP | J:244208 | |||||||||
| Biological Process | GO:0050915 | sensory perception of sour taste | ISO | J:164563 | |||||||||
| Biological Process | GO:0007224 | smoothened signaling pathway | IMP | J:207901 | |||||||||
| Biological Process | GO:0035725 | sodium ion transmembrane transport | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||