|
Symbol Name ID |
Nbn
nibrin MGI:1351625 |
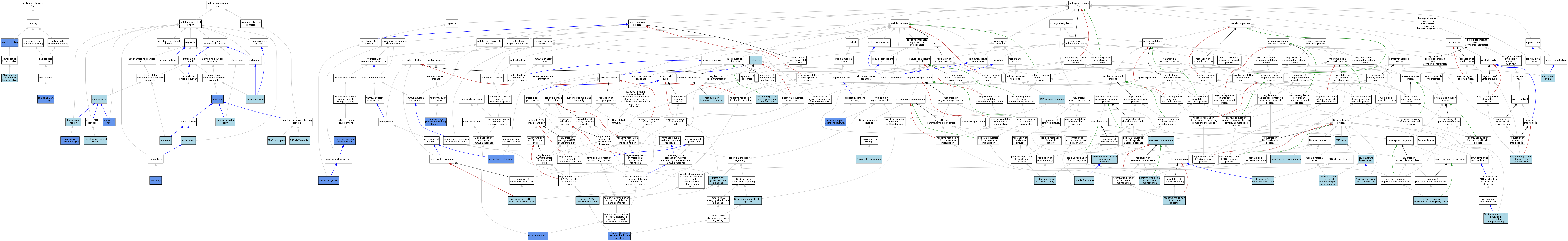
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003684 | damaged DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003684 | damaged DNA binding | IDA | J:76360 | |||||||||
| Molecular Function | GO:0140297 | DNA-binding transcription factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:129762 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:147247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:265834 | |||||||||
| Cellular Component | GO:0070533 | BRCA1-C complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0098687 | chromosomal region | NAS | J:320151 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0000781 | chromosome, telomeric region | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000781 | chromosome, telomeric region | IDA | J:228107 | |||||||||
| Cellular Component | GO:0000781 | chromosome, telomeric region | IDA | J:228121 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030870 | Mre11 complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030870 | Mre11 complex | ISO | J:73065 | |||||||||
| Cellular Component | GO:0042405 | nuclear inclusion body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | NAS | J:319979 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:71817 | |||||||||
| Cellular Component | GO:0016605 | PML body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016605 | PML body | IDA | J:228107 | |||||||||
| Cellular Component | GO:0005657 | replication fork | IDA | J:76360 | |||||||||
| Cellular Component | GO:0035861 | site of double-strand break | ISO | J:164563 | |||||||||
| Biological Process | GO:0001832 | blastocyst growth | IMP | J:86563 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0000077 | DNA damage checkpoint signaling | ISO | J:81636 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IEA | J:60000 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IEA | J:72247 | |||||||||
| Biological Process | GO:0000729 | DNA double-strand break processing | NAS | J:320151 | |||||||||
| Biological Process | GO:0032508 | DNA duplex unwinding | ISO | J:164563 | |||||||||
| Biological Process | GO:0032508 | DNA duplex unwinding | IBA | J:265628 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:72247 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:60000 | |||||||||
| Biological Process | GO:0110025 | DNA strand resection involved in replication fork processing | NAS | J:320083 | |||||||||
| Biological Process | GO:0006302 | double-strand break repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | IBA | J:265628 | |||||||||
| Biological Process | GO:0035825 | homologous recombination | NAS | J:319683 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | IMP | J:86563 | |||||||||
| Biological Process | GO:0097193 | intrinsic apoptotic signaling pathway | IMP | J:138550 | |||||||||
| Biological Process | GO:0045190 | isotype switching | IDA | J:96104 | |||||||||
| Biological Process | GO:0051321 | meiotic cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0007093 | mitotic cell cycle checkpoint signaling | IEA | J:72247 | |||||||||
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | IMP | J:138550 | |||||||||
| Biological Process | GO:0044818 | mitotic G2/M transition checkpoint | NAS | J:320080 | |||||||||
| Biological Process | GO:0045665 | negative regulation of neuron differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:1904354 | negative regulation of telomere capping | ISO | J:164563 | |||||||||
| Biological Process | GO:0046597 | negative regulation of viral entry into host cell | ISO | J:155856 | |||||||||
| Biological Process | GO:0007405 | neuroblast proliferation | IMP | J:98316 | |||||||||
| Biological Process | GO:0007405 | neuroblast proliferation | IMP | J:98316 | |||||||||
| Biological Process | GO:0007405 | neuroblast proliferation | IMP | J:98316 | |||||||||
| Biological Process | GO:0050885 | neuromuscular process controlling balance | IMP | J:98316 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0033674 | positive regulation of kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0031954 | positive regulation of protein autophosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0032206 | positive regulation of telomere maintenance | ISO | J:164563 | |||||||||
| Biological Process | GO:0048145 | regulation of fibroblast proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0090656 | t-circle formation | ISO | J:164563 | |||||||||
| Biological Process | GO:0000723 | telomere maintenance | ISO | J:164563 | |||||||||
| Biological Process | GO:0090737 | telomere maintenance via telomere trimming | ISO | J:164563 | |||||||||
| Biological Process | GO:0031860 | telomeric 3' overhang formation | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||