|
Symbol Name ID |
Tlx3
T cell leukemia, homeobox 3 MGI:1351209 |
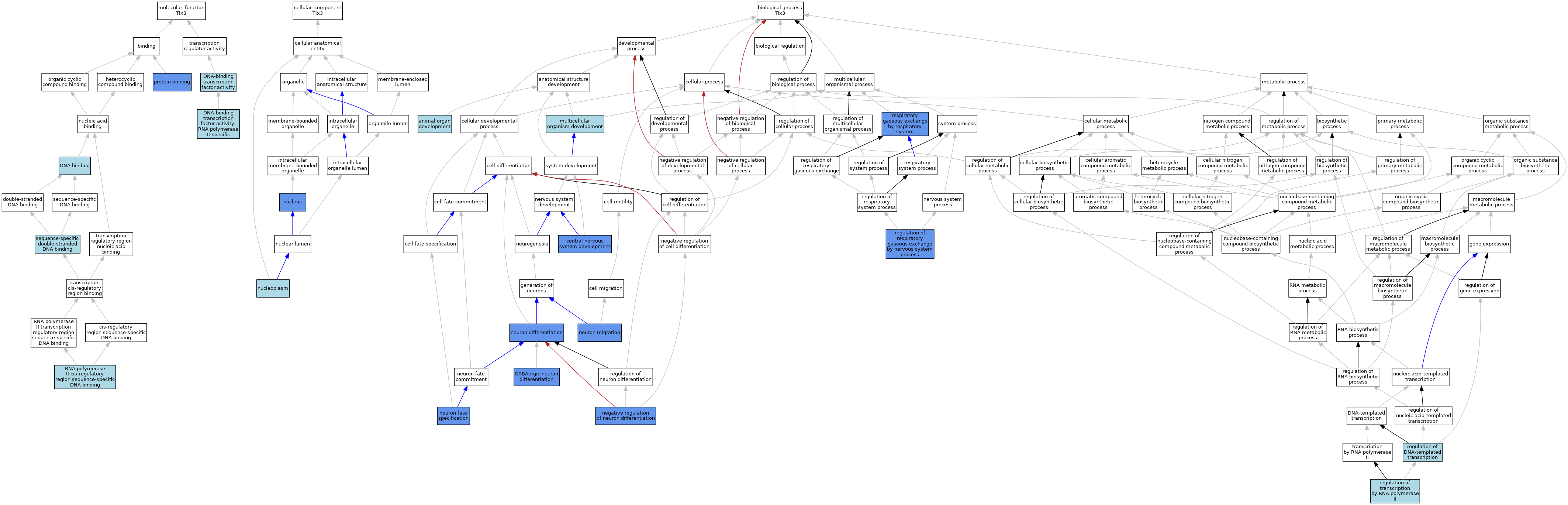
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:93761 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:93761 | |||||||||
| Biological Process | GO:0048513 | animal organ development | IBA | J:265628 | |||||||||
| Biological Process | GO:0007417 | central nervous system development | IMP | J:76655 | |||||||||
| Biological Process | GO:0097154 | GABAergic neuron differentiation | IGI | J:102513 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0045665 | negative regulation of neuron differentiation | IGI | J:102513 | |||||||||
| Biological Process | GO:0045665 | negative regulation of neuron differentiation | IGI | J:102513 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IMP | J:186470 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IMP | J:76655 | |||||||||
| Biological Process | GO:0048665 | neuron fate specification | IGI | J:90585 | |||||||||
| Biological Process | GO:0048665 | neuron fate specification | IMP | J:90585 | |||||||||
| Biological Process | GO:0048665 | neuron fate specification | IGI | J:102513 | |||||||||
| Biological Process | GO:0048665 | neuron fate specification | IGI | J:102513 | |||||||||
| Biological Process | GO:0001764 | neuron migration | IMP | J:92064 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
| Biological Process | GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | IMP | J:109165 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0007585 | respiratory gaseous exchange by respiratory system | IMP | J:60751 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||