|
Symbol Name ID |
Racgap1
Rac GTPase-activating protein 1 MGI:1349423 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
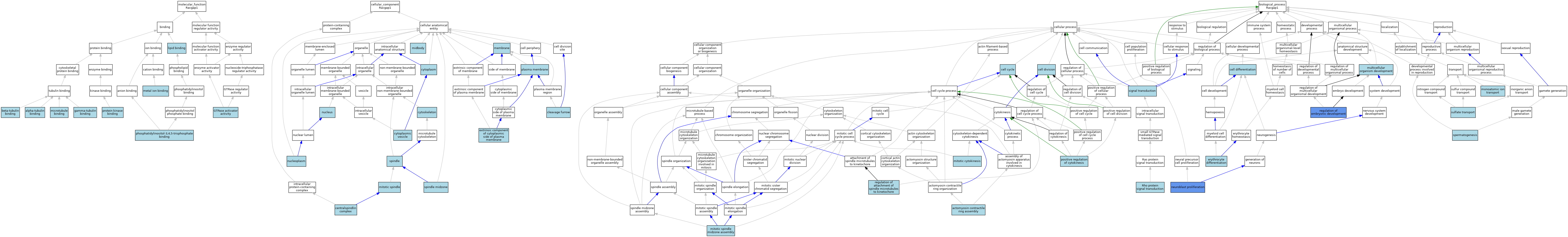
| Molecular Function | GO:0043014 | alpha-tubulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0048487 | beta-tubulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043015 | gamma-tubulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0097149 | centralspindlin complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0097149 | centralspindlin complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032154 | cleavage furrow | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032154 | cleavage furrow | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030496 | midbody | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030496 | midbody | IBA | J:265628 | |||||||||
| Cellular Component | GO:0072686 | mitotic spindle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005819 | spindle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0051233 | spindle midzone | ISO | J:164563 | |||||||||
| Cellular Component | GO:0051233 | spindle midzone | IBA | J:265628 | |||||||||
| Biological Process | GO:0000915 | actomyosin contractile ring assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0051301 | cell division | IEA | J:60000 | |||||||||
| Biological Process | GO:0030218 | erythrocyte differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0000281 | mitotic cytokinesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0000281 | mitotic cytokinesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0051256 | mitotic spindle midzone assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0051256 | mitotic spindle midzone assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0007405 | neuroblast proliferation | IEP | J:58013 | |||||||||
| Biological Process | GO:0032467 | positive regulation of cytokinesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | ISO | J:164563 | |||||||||
| Biological Process | GO:0045995 | regulation of embryonic development | IMP | J:69359 | |||||||||
| Biological Process | GO:0007266 | Rho protein signal transduction | IBA | J:265628 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IEA | J:60000 | |||||||||
| Biological Process | GO:0008272 | sulfate transport | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||