|
Symbol Name ID |
Foxg1
forkhead box G1 MGI:1347464 |
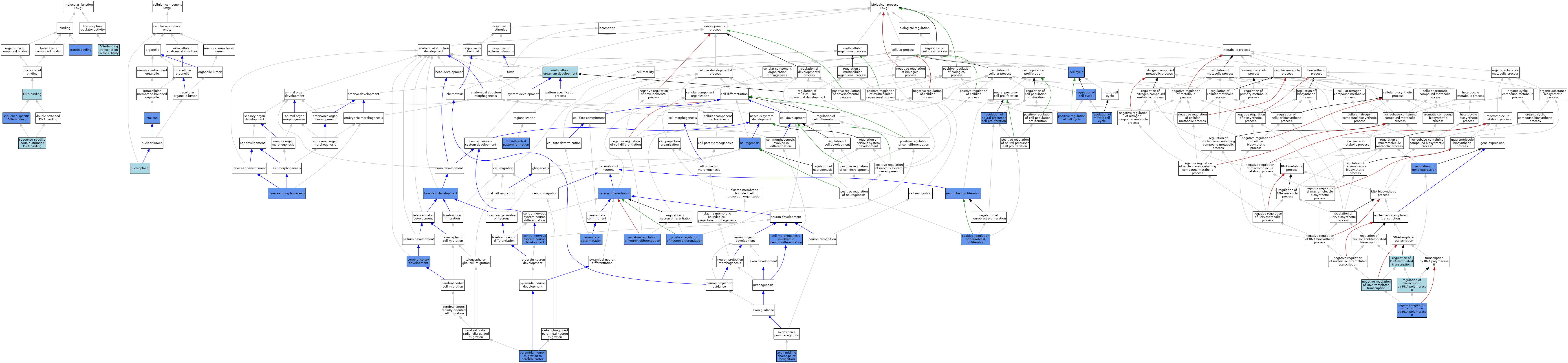
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:182500 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:235266 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:103764 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:103764 | |||||||||
| Molecular Function | GO:0043565 | sequence-specific DNA binding | IDA | J:241473 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9022604 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:141122 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:103764 | |||||||||
| Biological Process | GO:0016199 | axon midline choice point recognition | IMP | J:92062 | |||||||||
| Biological Process | GO:0016199 | axon midline choice point recognition | IMP | J:92062 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IMP | J:99329 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IMP | J:99329 | |||||||||
| Biological Process | GO:0048667 | cell morphogenesis involved in neuron differentiation | IMP | J:111604 | |||||||||
| Biological Process | GO:0021954 | central nervous system neuron development | IMP | J:110301 | |||||||||
| Biological Process | GO:0021987 | cerebral cortex development | IDA | J:188355 | |||||||||
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | IMP | J:86254 | |||||||||
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | IMP | J:99329 | |||||||||
| Biological Process | GO:0030900 | forebrain development | IMP | J:78115 | |||||||||
| Biological Process | GO:0030900 | forebrain development | IMP | J:87404 | |||||||||
| Biological Process | GO:0030900 | forebrain development | IMP | J:26720 | |||||||||
| Biological Process | GO:0030900 | forebrain development | IMP | J:86254 | |||||||||
| Biological Process | GO:0042472 | inner ear morphogenesis | IGI | J:111604 | |||||||||
| Biological Process | GO:0042472 | inner ear morphogenesis | IMP | J:111604 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0045665 | negative regulation of neuron differentiation | IMP | J:78115 | |||||||||
| Biological Process | GO:0045665 | negative regulation of neuron differentiation | IMP | J:87404 | |||||||||
| Biological Process | GO:0045665 | negative regulation of neuron differentiation | IMP | J:26720 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:103764 | |||||||||
| Biological Process | GO:0007405 | neuroblast proliferation | IMP | J:99329 | |||||||||
| Biological Process | GO:0007405 | neuroblast proliferation | IMP | J:99329 | |||||||||
| Biological Process | GO:0022008 | neurogenesis | IDA | J:133461 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IMP | J:26720 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IMP | J:87404 | |||||||||
| Biological Process | GO:0048664 | neuron fate determination | IDA | J:188355 | |||||||||
| Biological Process | GO:0045787 | positive regulation of cell cycle | IMP | J:99329 | |||||||||
| Biological Process | GO:0002052 | positive regulation of neuroblast proliferation | IMP | J:99329 | |||||||||
| Biological Process | GO:0045666 | positive regulation of neuron differentiation | IGI | J:103764 | |||||||||
| Biological Process | GO:0021852 | pyramidal neuron migration to cerebral cortex | IDA | J:188355 | |||||||||
| Biological Process | GO:0051726 | regulation of cell cycle | IDA | J:188355 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IMP | J:188355 | |||||||||
| Biological Process | GO:0007346 | regulation of mitotic cell cycle | IMP | J:78115 | |||||||||
| Biological Process | GO:2000177 | regulation of neural precursor cell proliferation | IGI | J:183998 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||