|
Symbol Name ID |
Ptpa
protein phosphatase 2 protein activator MGI:1346006 |
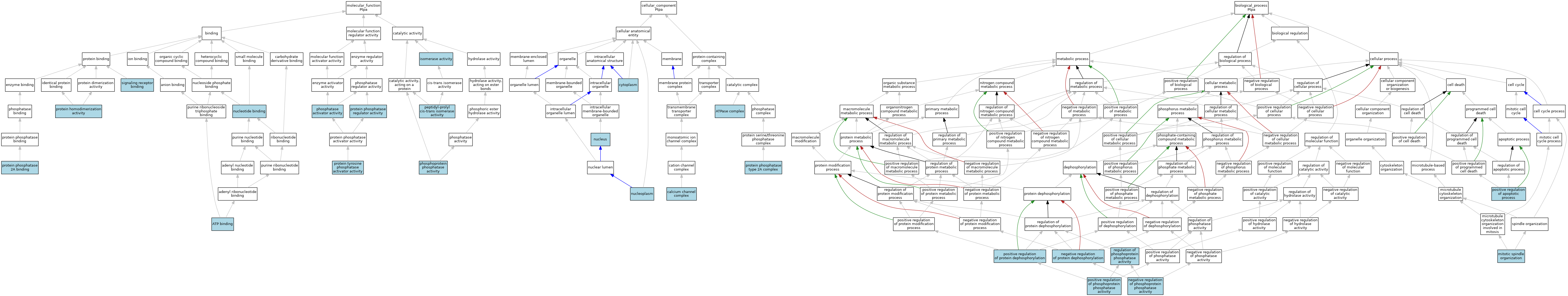
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016853 | isomerase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019211 | phosphatase activator activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004721 | phosphoprotein phosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051721 | protein phosphatase 2A binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019888 | protein phosphatase regulator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008160 | protein tyrosine phosphatase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008160 | protein tyrosine phosphatase activator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:1904949 | ATPase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0034704 | calcium channel complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000159 | protein phosphatase type 2A complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000159 | protein phosphatase type 2A complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0007052 | mitotic spindle organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0032515 | negative regulation of phosphoprotein phosphatase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0035308 | negative regulation of protein dephosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0035307 | positive regulation of protein dephosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043666 | regulation of phosphoprotein phosphatase activity | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||