|
Symbol Name ID |
Prkag2
protein kinase, AMP-activated, gamma 2 non-catalytic subunit MGI:1336153 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
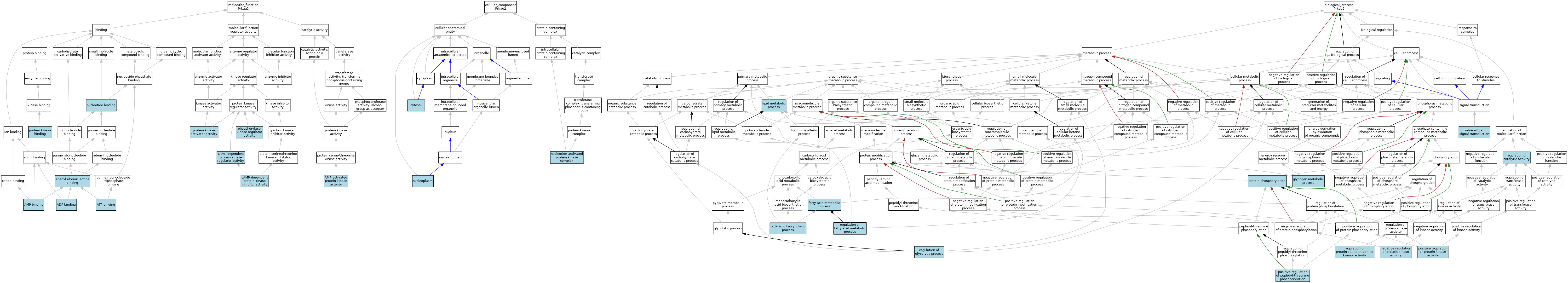
| Molecular Function | GO:0032559 | adenyl ribonucleotide binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0043531 | ADP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016208 | AMP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004679 | AMP-activated protein kinase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004679 | AMP-activated protein kinase activity | TAS | J:54164 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004862 | cAMP-dependent protein kinase inhibitor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008603 | cAMP-dependent protein kinase regulator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008607 | phosphorylase kinase regulator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030295 | protein kinase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-1454711 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9613545 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-1605475 | |||||||||
| Cellular Component | GO:0031588 | nucleotide-activated protein kinase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031588 | nucleotide-activated protein kinase complex | ISO | J:155856 | |||||||||
| Biological Process | GO:0006633 | fatty acid biosynthetic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006631 | fatty acid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0005977 | glycogen metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006469 | negative regulation of protein kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0045860 | positive regulation of protein kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0050790 | regulation of catalytic activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0019217 | regulation of fatty acid metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006110 | regulation of glycolytic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0071900 | regulation of protein serine/threonine kinase activity | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||