|
Symbol Name ID |
Zfpm2
zinc finger protein, multitype 2 MGI:1334444 |
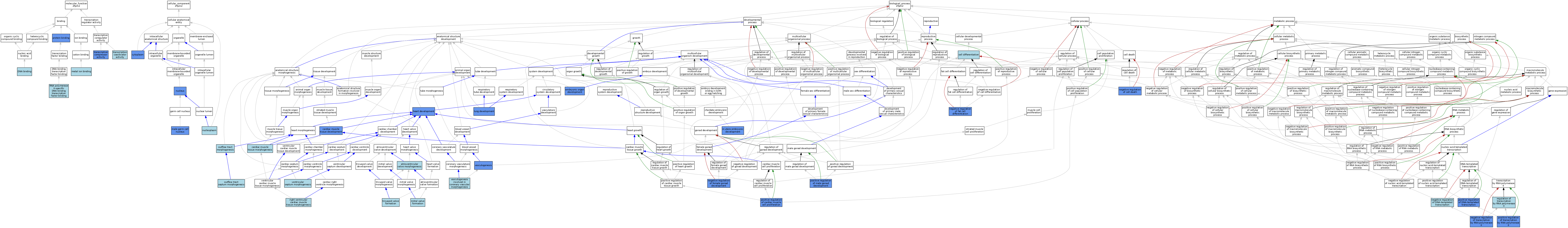
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:203454 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:172935 | |||||||||
| Molecular Function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IDA | J:53717 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IDA | J:82545 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:79154 | |||||||||
| Cellular Component | GO:0001673 | male germ cell nucleus | IDA | J:79154 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-9692234 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:172935 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:53717 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:82545 | |||||||||
| Biological Process | GO:0003181 | atrioventricular valve morphogenesis | TAS | J:99745 | |||||||||
| Biological Process | GO:0048738 | cardiac muscle tissue development | IMP | J:100119 | |||||||||
| Biological Process | GO:0055008 | cardiac muscle tissue morphogenesis | TAS | J:99745 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0048568 | embryonic organ development | ISO | J:100119 | |||||||||
| Biological Process | GO:0048568 | embryonic organ development | IMP | J:100119 | |||||||||
| Biological Process | GO:0007507 | heart development | IBA | J:265628 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:63121 | |||||||||
| Biological Process | GO:0007507 | heart development | IDA | J:62879 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:100119 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:100119 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:100119 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:100119 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | IMP | J:100119 | |||||||||
| Biological Process | GO:0030324 | lung development | IMP | J:100119 | |||||||||
| Biological Process | GO:0003192 | mitral valve formation | TAS | J:99745 | |||||||||
| Biological Process | GO:0060548 | negative regulation of cell death | IMP | J:172935 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | TAS | J:169941 | |||||||||
| Biological Process | GO:0045599 | negative regulation of fat cell differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0045599 | negative regulation of fat cell differentiation | IMP | J:172935 | |||||||||
| Biological Process | GO:2000195 | negative regulation of female gonad development | IMP | J:147038 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:82545 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:53717 | |||||||||
| Biological Process | GO:0003151 | outflow tract morphogenesis | TAS | J:99745 | |||||||||
| Biological Process | GO:0003148 | outflow tract septum morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0060045 | positive regulation of cardiac muscle cell proliferation | IGI | J:231272 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | TAS | J:169941 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IMP | J:147038 | |||||||||
| Biological Process | GO:2000020 | positive regulation of male gonad development | IMP | J:147038 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IGI | J:98265 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IGI | J:98265 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IEA | J:72247 | |||||||||
| Biological Process | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0003195 | tricuspid valve formation | TAS | J:99745 | |||||||||
| Biological Process | GO:0001570 | vasculogenesis | IDA | J:62879 | |||||||||
| Biological Process | GO:0001570 | vasculogenesis | IMP | J:100119 | |||||||||
| Biological Process | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis | TAS | J:169941 | |||||||||
| Biological Process | GO:0060412 | ventricular septum morphogenesis | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||