|
Symbol Name ID |
Ran
RAN, member RAS oncogene family MGI:1333112 |
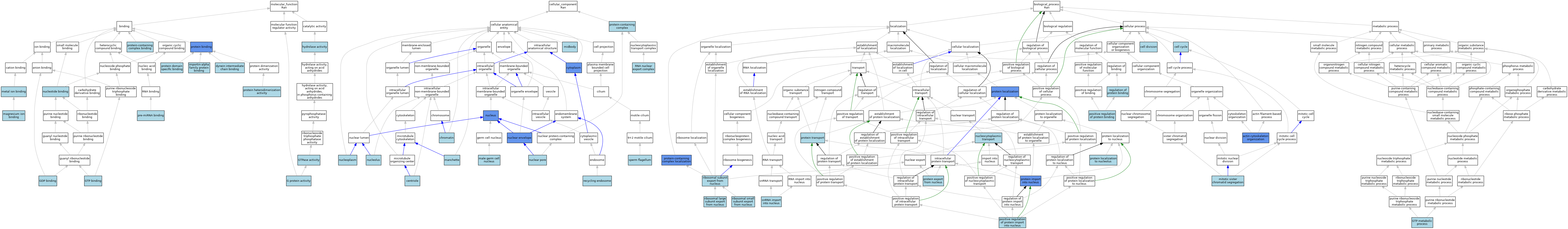
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0045505 | dynein intermediate chain binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003925 | G protein activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019003 | GDP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019003 | GDP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0061676 | importin-alpha family protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0000287 | magnesium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0070883 | pre-miRNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:222250 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:222250 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:74290 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:74290 | |||||||||
| Molecular Function | GO:0019904 | protein domain specific binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0046982 | protein heterodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005814 | centriole | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:137105 | |||||||||
| Cellular Component | GO:0001673 | male germ cell nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0002177 | manchette | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030496 | midbody | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005635 | nuclear envelope | IDA | J:222250 | |||||||||
| Cellular Component | GO:0005635 | nuclear envelope | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005643 | nuclear pore | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:222250 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:137105 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0055037 | recycling endosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042565 | RNA nuclear export complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0036126 | sperm flagellum | ISO | J:155856 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | IMP | J:176957 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0051301 | cell division | IEA | J:60000 | |||||||||
| Biological Process | GO:0046039 | GTP metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0000070 | mitotic sister chromatid segregation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006913 | nucleocytoplasmic transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0032092 | positive regulation of protein binding | ISO | J:164563 | |||||||||
| Biological Process | GO:0042307 | positive regulation of protein import into nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0006606 | protein import into nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0006606 | protein import into nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0006606 | protein import into nucleus | IDA | J:98705 | |||||||||
| Biological Process | GO:0006606 | protein import into nucleus | IDA | J:74290 | |||||||||
| Biological Process | GO:0008104 | protein localization | IMP | J:176957 | |||||||||
| Biological Process | GO:1902570 | protein localization to nucleolus | ISO | J:164563 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0031503 | protein-containing complex localization | IMP | J:176957 | |||||||||
| Biological Process | GO:0043393 | regulation of protein binding | ISO | J:155856 | |||||||||
| Biological Process | GO:0000055 | ribosomal large subunit export from nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0000056 | ribosomal small subunit export from nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0000054 | ribosomal subunit export from nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0061015 | snRNA import into nucleus | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||