|
Symbol Name ID |
Cldn3
claudin 3 MGI:1329044 |
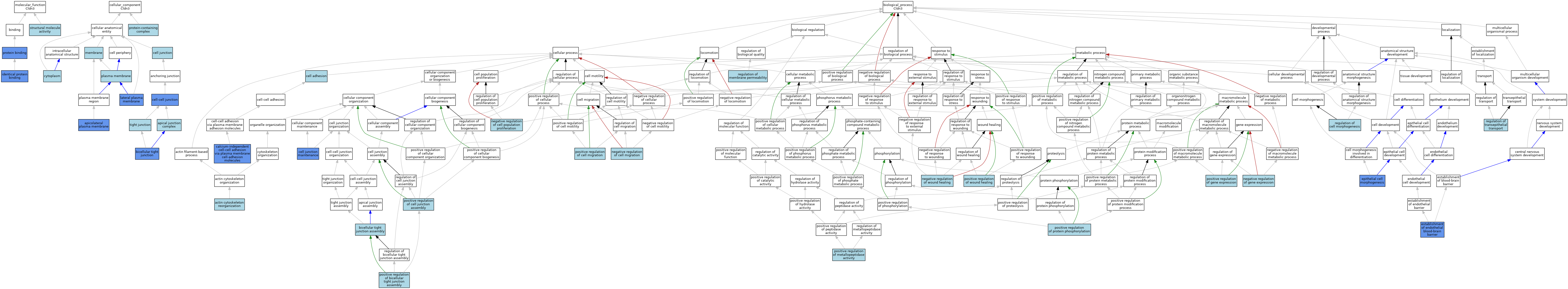
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:94758 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:58914 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:94758 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245332 | |||||||||
| Molecular Function | GO:0005198 | structural molecule activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0043296 | apical junction complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016327 | apicolateral plasma membrane | IDA | J:61744 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:61744 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:198884 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:181271 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:180511 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:180511 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:230002 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:107229 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:181271 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | IDA | J:280103 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016328 | lateral plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016328 | lateral plasma membrane | IDA | J:181271 | |||||||||
| Cellular Component | GO:0016020 | membrane | NAS | J:61744 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0070160 | tight junction | ISO | J:164563 | |||||||||
| Biological Process | GO:0031532 | actin cytoskeleton reorganization | ISO | J:164563 | |||||||||
| Biological Process | GO:0070830 | bicellular tight junction assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0070830 | bicellular tight junction assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | IDA | J:113790 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IBA | J:265628 | |||||||||
| Biological Process | GO:0034331 | cell junction maintenance | IMP | J:280103 | |||||||||
| Biological Process | GO:0003382 | epithelial cell morphogenesis | IDA | J:198884 | |||||||||
| Biological Process | GO:0014045 | establishment of endothelial blood-brain barrier | IMP | J:280103 | |||||||||
| Biological Process | GO:0030336 | negative regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:0061045 | negative regulation of wound healing | ISO | J:164563 | |||||||||
| Biological Process | GO:1903348 | positive regulation of bicellular tight junction assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:1901890 | positive regulation of cell junction assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:1905050 | positive regulation of metallopeptidase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0090303 | positive regulation of wound healing | ISO | J:164563 | |||||||||
| Biological Process | GO:0022604 | regulation of cell morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0090559 | regulation of membrane permeability | ISO | J:164563 | |||||||||
| Biological Process | GO:0150111 | regulation of transepithelial transport | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/09/2024 MGI 6.23 |

|
|
|
||